Gene
KWMTBOMO04752
Pre Gene Modal
BGIBMGA009882
Annotation
PREDICTED:_nitrogen_permease_regulator_2-like_protein_[Papilio_polytes]
Full name
GATOR complex protein NPRL2
Alternative Name
Nitrogen permease regulator 2-like protein
Location in the cell
Extracellular Reliability : 1.166 Nuclear Reliability : 1.819
Sequence
CDS
ATGGATGGGAATATGAAATATTACGAAGGATGTGGCCAAGAAGGGCCCATTCGTTGTATATTCCTGTGTGAATTTCACCCCACCGCTGGACCCAAAATAACTTGCCAAGTGCCAGAAAATTACATTTCGAAAGATATTTTCGATACTGTCAGCCACTACATCATACCGAAGGTTCAACTTCAGCGATGCACCCTAACAGTGACCCTTCTCGGTTCGAAGATCCTGGGTTTCCCAGTGAGGATCGACAATAAGAAGTACGCCCGCAACGCGTATTATTTTAATCTTTGCTTCGTGTGCGACGCGTGGGCCAGGACTGTTCACCTTGAGCCCTTAGTCAAAAAACTCACAGAGTATCTACTATCGATGGAGTTGGAGTCAGAGTGGTTGTCGAAGCAGTCGACGTCTGGTGACGCGAAGGACCTCGGCTGTCTGATGCGGCAGGTCATGCAGGACATCAACTGCAGGAGAATGTGCACGCTGACCGTTGGTACGACAACGACCCACCTGACCGTGGTCCGGGTGAACAGCGATCCGGCTCCCGTTAAAGATCACCAGGTCCCGGTCTTCTTGTACAGCAGACGGTCTTTTGTGACGGACCAATGGGACCTCACCACTAATCAGATACTCCCCTACATTGACGGCTTCAATCACGTTTCCAAGATCGCAGCCCTGACGGACGTTGAGATCAGTCTGGTACGAGCCTGCGTCCAGAACCTGGTGTACTACGGCGTGGTGACCCTTGTGCCCATCTTCCAGTACTGCGCGGTCTACAGCGCGACGCCGAAGCTAAGACAGCTGACAAGATGCGCAGGCCTTCAGAGACAGTGCGTCGAATTCTGTGCCAGATCTCCACGGCACTTGCCCAAAGTCAGCGATATATTCCGTATGTACGCCGGAATGTCGTATGGGAGTACCATCAAGGACCTCTGCAGGAGGATGAAGCCACAGGAACTAGCTATAAACGAGAGGAAGTTGGTGCTATTCGGAGTGCTCGAGGGTCTTATACGAAGGGTTTATAAGTATCCAATAACTTTGAACGACGAGACGGCGTCGCTTCGAAGCGACCACAGCCAGTCGGTAGCGAGGACGTACAACGGGCTCGTGTGCCTGGACGAGCTGTGCTGCCAGGCGGGTCTGTCCGCGTCGCAGCTCGACGACCAACTGGAGCGGGACACGAACGTCGCCTTCATCGTCAAGTGA
Protein
MDGNMKYYEGCGQEGPIRCIFLCEFHPTAGPKITCQVPENYISKDIFDTVSHYIIPKVQLQRCTLTVTLLGSKILGFPVRIDNKKYARNAYYFNLCFVCDAWARTVHLEPLVKKLTEYLLSMELESEWLSKQSTSGDAKDLGCLMRQVMQDINCRRMCTLTVGTTTTHLTVVRVNSDPAPVKDHQVPVFLYSRRSFVTDQWDLTTNQILPYIDGFNHVSKIAALTDVEISLVRACVQNLVYYGVVTLVPIFQYCAVYSATPKLRQLTRCAGLQRQCVEFCARSPRHLPKVSDIFRMYAGMSYGSTIKDLCRRMKPQELAINERKLVLFGVLEGLIRRVYKYPITLNDETASLRSDHSQSVARTYNGLVCLDELCCQAGLSASQLDDQLERDTNVAFIVK
Summary
Description
An essential component of the GATOR subcomplex GATOR1 which functions as an inhibitor of the amino acid-sensing branch of the TORC1 signaling pathway (PubMed:23723238, PubMed:27166823, PubMed:25512509). The two GATOR subcomplexes, GATOR1 and GATOR2, regulate the TORC1 pathway in order to mediate metabolic homeostasis, female gametogenesis and the response to amino acid limitation and complete starvation (PubMed:23723238, PubMed:27166823, PubMed:25512509). The function of GATOR1 in negatively regulating the TORC1 pathway is essential for maintaining baseline levels of TORC1 activity under nutrient rich conditions, and for promoting survival during amino acid or complete starvation by inhibiting TORC1-dependent cell growth and promoting catabolic metabolism and autophagy (PubMed:23723238, PubMed:27166823). In addition, this inhibition of TORC1 is necessary to maintain female fertility under normal conditions and during periods of nutrient stress (PubMed:24786828, PubMed:27672113, PubMed:25512509). GATOR1 and GATOR2 act at different stages of oogenesis to regulate TORC1 in order to control meiotic entry and promote oocyte growth and development (PubMed:25512509). After exactly four mitotic cyst divisions, the GATOR1 complex members (Iml1, Nprl2 and Nprl3) down-regulate TORC1 to slow cellular metabolism and promote the mitotic/meiotic transition (PubMed:25512509). At later stages of oogenesis, the mio and Nup44A components of the GATOR2 complex inhibit GATOR1 and thus activate TORC1 to promote meiotic progression, and drive oocyte growth and development (PubMed:25512509).
Subunit
Component of the GATOR complex consisting of mio, Nup44A/Seh1, Im11, Nplr3, Nplr2, Wdr24, Wdr59 and Sec13 (PubMed:27166823). Within the GATOR complex, probable component of the GATOR1 subcomplex which is likely composed of Iml1, Nplr2 and Nplr3 (PubMed:27166823). Interacts with Nprl3 (PubMed:24786828).
Similarity
Belongs to the cyclin family.
Belongs to the NPR2 family.
Belongs to the NPR2 family.
Keywords
Cell cycle
Cell division
Complete proteome
Cytoplasm
Lysosome
Meiosis
Mitosis
Reference proteome
Feature
chain GATOR complex protein NPRL2
Uniprot
A0A0N1I4N6
A0A194PRZ5
A0A067QLB4
K7ITL1
A0A310SKD5
A0A2A3EM38
+ More
A0A087ZVT4 E2ACA6 E2BDB2 A0A154PQN0 A0A1B6CWT5 A0A151IFG8 A0A2J7PXT5 A0A151XAD9 A0A0L7QK55 A0A195B7V6 A0A158NGR1 A0A1B6MCH0 E9ILD1 A0A195FFE7 A0A026WPQ6 A0A195DYG6 F4WZK8 A0A0N0BEH8 A0A1W4X2D4 A0A3L8DMV7 A0A1B6J7B9 A0A0T6B7V0 A0A0V0GBZ7 A0A023F0H1 A0A224XDW6 T1HAA8 A0A1Y1N970 D6WVQ8 A0A069DTW7 A0A0A9Y5U1 J3JYJ9 A0A067R7Y0 E9HBY6 A0A0P6AWU2 A0A0P5IXD7 A0A182H4E0 A0A182H3M3 Q17EA9 A0A1S4G401 Q16E62 W8BQH1 A0A1A9YF26 A0A1A9VUK7 A0A1B0AMR6 A0A1B0A0V7 E0V9K4 A0A1I8Q9U5 U5EZU2 A0A1I8MUY4 B0XL24 B4Q1W7 B4IEU6 B4M2A5 A0A232F856 B4R647 B3NW96 A0A0K8UFV4 Q9VXA0 A0A034WGG6 A0A3B0J7B3 Q29FL0 B4HA42 A0A0A1XIV2 B4L1V0 A0A1B6GSQ5 A0A1W4VIY6 B4NCB0 B3N0C1 A0A1Z5L6Y0 B4JL56 T1IUN9 A0A0M3QYV4 A0A0P5MXS4 A0A162QCX1 A0A1A9W648 A0A1S3HQR9 A0A0P5X0K2 A0A182QPI2 A0A0P5KB23 A0A0K8TMA4 A0A182XXU5 A0A224YHX9 A0A1W4YNM8 A0A131Z4R3 A0A3S3RSE4 A0A182VMS5 A0A182XDT4 Q7QFE3 A0A182IAS0 A0A182U1G1 A0A182W5G9 A0A182N1A1
A0A087ZVT4 E2ACA6 E2BDB2 A0A154PQN0 A0A1B6CWT5 A0A151IFG8 A0A2J7PXT5 A0A151XAD9 A0A0L7QK55 A0A195B7V6 A0A158NGR1 A0A1B6MCH0 E9ILD1 A0A195FFE7 A0A026WPQ6 A0A195DYG6 F4WZK8 A0A0N0BEH8 A0A1W4X2D4 A0A3L8DMV7 A0A1B6J7B9 A0A0T6B7V0 A0A0V0GBZ7 A0A023F0H1 A0A224XDW6 T1HAA8 A0A1Y1N970 D6WVQ8 A0A069DTW7 A0A0A9Y5U1 J3JYJ9 A0A067R7Y0 E9HBY6 A0A0P6AWU2 A0A0P5IXD7 A0A182H4E0 A0A182H3M3 Q17EA9 A0A1S4G401 Q16E62 W8BQH1 A0A1A9YF26 A0A1A9VUK7 A0A1B0AMR6 A0A1B0A0V7 E0V9K4 A0A1I8Q9U5 U5EZU2 A0A1I8MUY4 B0XL24 B4Q1W7 B4IEU6 B4M2A5 A0A232F856 B4R647 B3NW96 A0A0K8UFV4 Q9VXA0 A0A034WGG6 A0A3B0J7B3 Q29FL0 B4HA42 A0A0A1XIV2 B4L1V0 A0A1B6GSQ5 A0A1W4VIY6 B4NCB0 B3N0C1 A0A1Z5L6Y0 B4JL56 T1IUN9 A0A0M3QYV4 A0A0P5MXS4 A0A162QCX1 A0A1A9W648 A0A1S3HQR9 A0A0P5X0K2 A0A182QPI2 A0A0P5KB23 A0A0K8TMA4 A0A182XXU5 A0A224YHX9 A0A1W4YNM8 A0A131Z4R3 A0A3S3RSE4 A0A182VMS5 A0A182XDT4 Q7QFE3 A0A182IAS0 A0A182U1G1 A0A182W5G9 A0A182N1A1
Pubmed
26354079
24845553
20075255
20798317
21347285
21282665
+ More
24508170 21719571 30249741 25474469 28004739 18362917 19820115 26334808 25401762 26823975 22516182 23537049 21292972 26483478 17510324 24495485 20566863 25315136 17994087 17550304 28648823 10731132 12537572 12537569 23723238 24786828 25512509 27672113 27166823 25348373 15632085 25830018 28528879 26369729 25244985 28797301 26830274 12364791 14747013 17210077
24508170 21719571 30249741 25474469 28004739 18362917 19820115 26334808 25401762 26823975 22516182 23537049 21292972 26483478 17510324 24495485 20566863 25315136 17994087 17550304 28648823 10731132 12537572 12537569 23723238 24786828 25512509 27672113 27166823 25348373 15632085 25830018 28528879 26369729 25244985 28797301 26830274 12364791 14747013 17210077
EMBL
KQ461192
KPJ07023.1
KQ459595
KPI95728.1
KK853297
KDR08783.1
+ More
KQ763450 OAD55051.1 KZ288219 PBC32229.1 GL438491 EFN68896.1 GL447585 EFN86340.1 KQ435048 KZC14209.1 GEDC01019358 JAS17940.1 KQ977793 KYM99659.1 NEVH01020852 PNF21147.1 KQ982339 KYQ57322.1 KQ415041 KOC58901.1 KQ976574 KYM80279.1 ADTU01015170 GEBQ01006360 JAT33617.1 GL764074 EFZ18599.1 KQ981625 KYN39081.1 KK107135 EZA58042.1 KQ980107 KYN17649.1 GL888480 EGI60283.1 KQ435833 KOX71614.1 QOIP01000006 RLU21711.1 GECU01012639 JAS95067.1 LJIG01009518 KRT82919.1 GECL01000597 JAP05527.1 GBBI01003762 JAC14950.1 GFTR01005781 JAW10645.1 ACPB03021733 GEZM01013053 JAV92796.1 KQ971358 EFA08261.1 GBGD01001752 JAC87137.1 GBHO01015172 GDHC01006029 JAG28432.1 JAQ12600.1 APGK01056842 BT128328 KB741277 KB632028 AEE63286.1 ENN71159.1 ERL88120.1 KK852819 KDR15634.1 GL732618 EFX70671.1 GDIP01023243 JAM80472.1 GDIQ01208454 GDIQ01208453 JAK43272.1 JXUM01109210 KQ565299 KXJ71113.1 JXUM01024583 KQ560695 KXJ81235.1 CH477284 EAT44769.1 CH479067 EAT32519.1 GAMC01007492 JAB99063.1 JXJN01000553 JXJN01000554 JXJN01000555 DS234994 EEB10060.1 GANO01000142 JAB59729.1 DS234210 EDS33167.1 CM000162 EDX02542.1 CH480832 EDW46200.1 CH940651 EDW65809.1 NNAY01000761 OXU26639.1 CM000366 EDX18151.1 CH954180 EDV46435.1 GDHF01027109 JAI25205.1 AE003503 AY051914 AAK93338.1 GAKP01005748 GAKP01005747 JAC53205.1 OUUW01000003 SPP78124.1 CH379066 EAL31390.1 CH479235 EDW36710.1 GBXI01003415 JAD10877.1 CH933810 EDW06753.1 GECZ01004308 JAS65461.1 CH964239 EDW82469.1 CH902640 EDV38325.1 GFJQ02003814 JAW03156.1 CH916370 EDW00309.1 JH431551 CP012528 ALC48269.1 GDIQ01151969 JAK99756.1 LRGB01000359 KZS19577.1 GDIP01079402 JAM24313.1 AXCN02001237 GDIQ01212706 JAK39019.1 GDAI01002106 JAI15497.1 GFPF01005429 MAA16575.1 GEDV01002589 JAP85968.1 NCKU01005917 NCKU01005916 RWS04038.1 RWS04041.1 AAAB01008846 EAA06517.3 APCN01001313
KQ763450 OAD55051.1 KZ288219 PBC32229.1 GL438491 EFN68896.1 GL447585 EFN86340.1 KQ435048 KZC14209.1 GEDC01019358 JAS17940.1 KQ977793 KYM99659.1 NEVH01020852 PNF21147.1 KQ982339 KYQ57322.1 KQ415041 KOC58901.1 KQ976574 KYM80279.1 ADTU01015170 GEBQ01006360 JAT33617.1 GL764074 EFZ18599.1 KQ981625 KYN39081.1 KK107135 EZA58042.1 KQ980107 KYN17649.1 GL888480 EGI60283.1 KQ435833 KOX71614.1 QOIP01000006 RLU21711.1 GECU01012639 JAS95067.1 LJIG01009518 KRT82919.1 GECL01000597 JAP05527.1 GBBI01003762 JAC14950.1 GFTR01005781 JAW10645.1 ACPB03021733 GEZM01013053 JAV92796.1 KQ971358 EFA08261.1 GBGD01001752 JAC87137.1 GBHO01015172 GDHC01006029 JAG28432.1 JAQ12600.1 APGK01056842 BT128328 KB741277 KB632028 AEE63286.1 ENN71159.1 ERL88120.1 KK852819 KDR15634.1 GL732618 EFX70671.1 GDIP01023243 JAM80472.1 GDIQ01208454 GDIQ01208453 JAK43272.1 JXUM01109210 KQ565299 KXJ71113.1 JXUM01024583 KQ560695 KXJ81235.1 CH477284 EAT44769.1 CH479067 EAT32519.1 GAMC01007492 JAB99063.1 JXJN01000553 JXJN01000554 JXJN01000555 DS234994 EEB10060.1 GANO01000142 JAB59729.1 DS234210 EDS33167.1 CM000162 EDX02542.1 CH480832 EDW46200.1 CH940651 EDW65809.1 NNAY01000761 OXU26639.1 CM000366 EDX18151.1 CH954180 EDV46435.1 GDHF01027109 JAI25205.1 AE003503 AY051914 AAK93338.1 GAKP01005748 GAKP01005747 JAC53205.1 OUUW01000003 SPP78124.1 CH379066 EAL31390.1 CH479235 EDW36710.1 GBXI01003415 JAD10877.1 CH933810 EDW06753.1 GECZ01004308 JAS65461.1 CH964239 EDW82469.1 CH902640 EDV38325.1 GFJQ02003814 JAW03156.1 CH916370 EDW00309.1 JH431551 CP012528 ALC48269.1 GDIQ01151969 JAK99756.1 LRGB01000359 KZS19577.1 GDIP01079402 JAM24313.1 AXCN02001237 GDIQ01212706 JAK39019.1 GDAI01002106 JAI15497.1 GFPF01005429 MAA16575.1 GEDV01002589 JAP85968.1 NCKU01005917 NCKU01005916 RWS04038.1 RWS04041.1 AAAB01008846 EAA06517.3 APCN01001313
Proteomes
UP000053240
UP000053268
UP000027135
UP000002358
UP000242457
UP000005203
+ More
UP000000311 UP000008237 UP000076502 UP000078542 UP000235965 UP000075809 UP000053825 UP000078540 UP000005205 UP000078541 UP000053097 UP000078492 UP000007755 UP000053105 UP000192223 UP000279307 UP000015103 UP000007266 UP000019118 UP000030742 UP000000305 UP000069940 UP000249989 UP000008820 UP000092443 UP000078200 UP000092460 UP000092445 UP000009046 UP000095300 UP000095301 UP000002320 UP000002282 UP000001292 UP000008792 UP000215335 UP000000304 UP000008711 UP000000803 UP000268350 UP000001819 UP000008744 UP000009192 UP000192221 UP000007798 UP000007801 UP000001070 UP000092553 UP000076858 UP000091820 UP000085678 UP000075886 UP000076408 UP000192224 UP000285301 UP000075903 UP000076407 UP000007062 UP000075840 UP000075902 UP000075920 UP000075884
UP000000311 UP000008237 UP000076502 UP000078542 UP000235965 UP000075809 UP000053825 UP000078540 UP000005205 UP000078541 UP000053097 UP000078492 UP000007755 UP000053105 UP000192223 UP000279307 UP000015103 UP000007266 UP000019118 UP000030742 UP000000305 UP000069940 UP000249989 UP000008820 UP000092443 UP000078200 UP000092460 UP000092445 UP000009046 UP000095300 UP000095301 UP000002320 UP000002282 UP000001292 UP000008792 UP000215335 UP000000304 UP000008711 UP000000803 UP000268350 UP000001819 UP000008744 UP000009192 UP000192221 UP000007798 UP000007801 UP000001070 UP000092553 UP000076858 UP000091820 UP000085678 UP000075886 UP000076408 UP000192224 UP000285301 UP000075903 UP000076407 UP000007062 UP000075840 UP000075902 UP000075920 UP000075884
PRIDE
Interpro
IPR009348
NPR2
+ More
IPR036188 FAD/NAD-bd_sf
IPR023753 FAD/NAD-binding_dom
IPR013763 Cyclin-like
IPR006671 Cyclin_N
IPR015904 Sulphide_quinone_reductase
IPR036915 Cyclin-like_sf
IPR008936 Rho_GTPase_activation_prot
IPR000198 RhoGAP_dom
IPR029071 Ubiquitin-like_domsf
IPR001849 PH_domain
IPR000159 RA_dom
IPR036188 FAD/NAD-bd_sf
IPR023753 FAD/NAD-binding_dom
IPR013763 Cyclin-like
IPR006671 Cyclin_N
IPR015904 Sulphide_quinone_reductase
IPR036915 Cyclin-like_sf
IPR008936 Rho_GTPase_activation_prot
IPR000198 RhoGAP_dom
IPR029071 Ubiquitin-like_domsf
IPR001849 PH_domain
IPR000159 RA_dom
Gene 3D
CDD
ProteinModelPortal
A0A0N1I4N6
A0A194PRZ5
A0A067QLB4
K7ITL1
A0A310SKD5
A0A2A3EM38
+ More
A0A087ZVT4 E2ACA6 E2BDB2 A0A154PQN0 A0A1B6CWT5 A0A151IFG8 A0A2J7PXT5 A0A151XAD9 A0A0L7QK55 A0A195B7V6 A0A158NGR1 A0A1B6MCH0 E9ILD1 A0A195FFE7 A0A026WPQ6 A0A195DYG6 F4WZK8 A0A0N0BEH8 A0A1W4X2D4 A0A3L8DMV7 A0A1B6J7B9 A0A0T6B7V0 A0A0V0GBZ7 A0A023F0H1 A0A224XDW6 T1HAA8 A0A1Y1N970 D6WVQ8 A0A069DTW7 A0A0A9Y5U1 J3JYJ9 A0A067R7Y0 E9HBY6 A0A0P6AWU2 A0A0P5IXD7 A0A182H4E0 A0A182H3M3 Q17EA9 A0A1S4G401 Q16E62 W8BQH1 A0A1A9YF26 A0A1A9VUK7 A0A1B0AMR6 A0A1B0A0V7 E0V9K4 A0A1I8Q9U5 U5EZU2 A0A1I8MUY4 B0XL24 B4Q1W7 B4IEU6 B4M2A5 A0A232F856 B4R647 B3NW96 A0A0K8UFV4 Q9VXA0 A0A034WGG6 A0A3B0J7B3 Q29FL0 B4HA42 A0A0A1XIV2 B4L1V0 A0A1B6GSQ5 A0A1W4VIY6 B4NCB0 B3N0C1 A0A1Z5L6Y0 B4JL56 T1IUN9 A0A0M3QYV4 A0A0P5MXS4 A0A162QCX1 A0A1A9W648 A0A1S3HQR9 A0A0P5X0K2 A0A182QPI2 A0A0P5KB23 A0A0K8TMA4 A0A182XXU5 A0A224YHX9 A0A1W4YNM8 A0A131Z4R3 A0A3S3RSE4 A0A182VMS5 A0A182XDT4 Q7QFE3 A0A182IAS0 A0A182U1G1 A0A182W5G9 A0A182N1A1
A0A087ZVT4 E2ACA6 E2BDB2 A0A154PQN0 A0A1B6CWT5 A0A151IFG8 A0A2J7PXT5 A0A151XAD9 A0A0L7QK55 A0A195B7V6 A0A158NGR1 A0A1B6MCH0 E9ILD1 A0A195FFE7 A0A026WPQ6 A0A195DYG6 F4WZK8 A0A0N0BEH8 A0A1W4X2D4 A0A3L8DMV7 A0A1B6J7B9 A0A0T6B7V0 A0A0V0GBZ7 A0A023F0H1 A0A224XDW6 T1HAA8 A0A1Y1N970 D6WVQ8 A0A069DTW7 A0A0A9Y5U1 J3JYJ9 A0A067R7Y0 E9HBY6 A0A0P6AWU2 A0A0P5IXD7 A0A182H4E0 A0A182H3M3 Q17EA9 A0A1S4G401 Q16E62 W8BQH1 A0A1A9YF26 A0A1A9VUK7 A0A1B0AMR6 A0A1B0A0V7 E0V9K4 A0A1I8Q9U5 U5EZU2 A0A1I8MUY4 B0XL24 B4Q1W7 B4IEU6 B4M2A5 A0A232F856 B4R647 B3NW96 A0A0K8UFV4 Q9VXA0 A0A034WGG6 A0A3B0J7B3 Q29FL0 B4HA42 A0A0A1XIV2 B4L1V0 A0A1B6GSQ5 A0A1W4VIY6 B4NCB0 B3N0C1 A0A1Z5L6Y0 B4JL56 T1IUN9 A0A0M3QYV4 A0A0P5MXS4 A0A162QCX1 A0A1A9W648 A0A1S3HQR9 A0A0P5X0K2 A0A182QPI2 A0A0P5KB23 A0A0K8TMA4 A0A182XXU5 A0A224YHX9 A0A1W4YNM8 A0A131Z4R3 A0A3S3RSE4 A0A182VMS5 A0A182XDT4 Q7QFE3 A0A182IAS0 A0A182U1G1 A0A182W5G9 A0A182N1A1
PDB
6CET
E-value=9.67738e-82,
Score=773
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Localizes primarily to the autolysosomes during amino-acid starvation. With evidence from 2 publications.
Lysosome Localizes primarily to the autolysosomes during amino-acid starvation. With evidence from 2 publications.
Lysosome Localizes primarily to the autolysosomes during amino-acid starvation. With evidence from 2 publications.
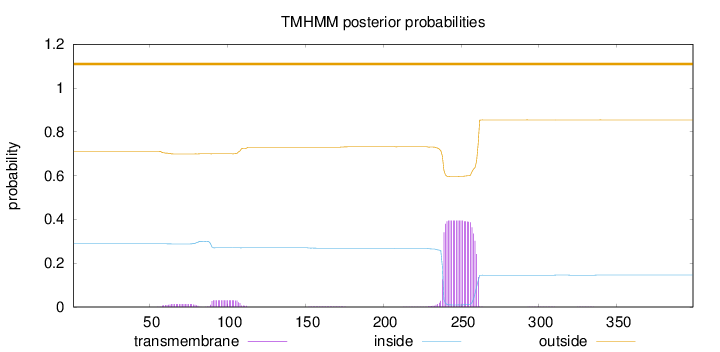
Length:
399
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.52240999999998
Exp number, first 60 AAs:
0.02274
Total prob of N-in:
0.28878
outside
1 - 399
Population Genetic Test Statistics
Pi
260.155429
Theta
156.931425
Tajima's D
1.954686
CLR
0.247586
CSRT
0.875506224688766
Interpretation
Uncertain
