Gene
KWMTBOMO04751
Pre Gene Modal
BGIBMGA009859
Annotation
PREDICTED:_phosphopantothenoylcysteine_decarboxylase_[Papilio_polytes]
Full name
Phosphopantothenoylcysteine decarboxylase
Alternative Name
CoaC
Location in the cell
Cytoplasmic Reliability : 2.151
Sequence
CDS
ATGGATGAAAATACTACATTTAATGTTTTAGTAGGTGTTACTGGAAGTGTTGCAGCTATAAAGATACCGAATTTAATAAAATCCGTGTTGGAGTTACGAAGCGAGACTCGATATAATTTTAAGGTACGACTAATAGTAACAGAACATGCAAAGCACTTCTTTGAGTGGACTGAAATGCCCTTGGATGTACTAAGATATGATGACTCTTCAGAATGGGTAACATGGAAAGGAAGAGGAGATCCAGTATTACATATAGAACTTGGAAAATGGGCTGACATAATGGTTATAGCACCAATGGATGCTAATACATTAGCAAAAGTGGCATTGGGCATGTGTGATAATTTAGTCACATGTACAGTAAGAGCATGGAACTTCTCCAAACCTCTGCTATTTTGTCCAGCTATGAATACTAGAATGTGGGACCATCCAATCACAGCCACTCACATTTCTACACTACGAGCCTGGGGTTTTATTCAAATACCACCCATAACTAAAACTTTGATGTGCGGTGACACCGGTGTAGGTGCCATGGCAGAAGTAGAGGCAATTGTTGAAAAAATCAAATTAATAGCTGATATGAAAATGAATAGAACACAATAA
Protein
MDENTTFNVLVGVTGSVAAIKIPNLIKSVLELRSETRYNFKVRLIVTEHAKHFFEWTEMPLDVLRYDDSSEWVTWKGRGDPVLHIELGKWADIMVIAPMDANTLAKVALGMCDNLVTCTVRAWNFSKPLLFCPAMNTRMWDHPITATHISTLRAWGFIQIPPITKTLMCGDTGVGAMAEVEAIVEKIKLIADMKMNRTQ
Summary
Description
Necessary for the biosynthesis of coenzyme A. Catalyzes the decarboxylation of 4-phosphopantothenoylcysteine to form 4'-phosphopantotheine (By similarity).
Catalytic Activity
H(+) + N-[(R)-4-phosphopantothenoyl]-L-cysteine = CO2 + D-pantetheine 4'-phosphate
Cofactor
FMN
Subunit
Homotrimer.
Similarity
Belongs to the HFCD (homooligomeric flavin containing Cys decarboxylase) superfamily.
Keywords
Coenzyme A biosynthesis
Complete proteome
Decarboxylase
Flavoprotein
FMN
Lyase
Reference proteome
Feature
chain Phosphopantothenoylcysteine decarboxylase
Uniprot
H9JK08
A0A0N0PAJ4
A0A2H1WRW2
A0A194PSJ4
A0A2A4JEQ2
A0A212FCL2
+ More
A0A182KBI3 A0A2J7QP62 A0A2M4AWQ4 A0A0L0CQP0 Q7PZN2 A0A182XFQ3 A0A182FVS7 A0A2M3ZCL9 A0A182UQE4 A0A182HHS1 A0A2M3ZJK1 U5ESC8 A0A2M4C078 A0A1I8NPL5 A0A182KLL6 A0A067QIV1 A0A182W255 A0A2M3ZCS2 A0A084W4E2 A0A023EIV9 A0A034WTA2 A0A1I8MJB3 D2CFX2 A0A0K8USB0 A0A0K8TKK5 A0A182MV92 W8BHG4 W5JPY6 Q1HQE9 A0A182N9P8 A0A182RMQ5 A0A0L7L7Q6 J3JV50 A0A1Y1KW16 A0A1W4WXV5 U4TS01 A0A182R154 B0XL28 A0A1S4ETD4 A0A182J918 A0A3B3Z9U4 A0A0M3QUP7 A0A1Q3F7Z7 I3KE39 A0A250XW22 U3K8G5 A0A1B6FE18 A0A3B3Z9A2 A0A3P9D6X9 A0A1A9W118 A0A3Q4MXS0 H9GP06 A0A1Q3FIK7 A0A091RTN1 A0A1Q3FI41 A0A1Q3FJC3 A0A091KDT5 A0A3P8Q5B4 A0A3P9D7J4 A0A091LNL9 G1MTN7 A0A1A9XZK9 A0A091TWZ1 D3TRH5 A0A3L8RUI6 V5G7P2 A0A1B0G7E8 H0Z086 A0A091NJ21 A0A3Q4MXQ3 G3U899 E3TGC7 A0A0A0AM39 Q6PBQ5 Q1LWK2 K7FZ56 B4JW85 A0A091MGQ1 B0S701 B4P8F9 A0A093JBD5 A0A3P8PME1 A0A1U8BLH5 Q8BZB2 A0A3B4H8R4 I3MDG5 E1BUI2 A0A091P7J7 V9L0R4 A0A1B6JRG1 A0A2R7WHC0 A0A3Q1CJ39 A0A2Y9QQ67 A0A093EFJ3
A0A182KBI3 A0A2J7QP62 A0A2M4AWQ4 A0A0L0CQP0 Q7PZN2 A0A182XFQ3 A0A182FVS7 A0A2M3ZCL9 A0A182UQE4 A0A182HHS1 A0A2M3ZJK1 U5ESC8 A0A2M4C078 A0A1I8NPL5 A0A182KLL6 A0A067QIV1 A0A182W255 A0A2M3ZCS2 A0A084W4E2 A0A023EIV9 A0A034WTA2 A0A1I8MJB3 D2CFX2 A0A0K8USB0 A0A0K8TKK5 A0A182MV92 W8BHG4 W5JPY6 Q1HQE9 A0A182N9P8 A0A182RMQ5 A0A0L7L7Q6 J3JV50 A0A1Y1KW16 A0A1W4WXV5 U4TS01 A0A182R154 B0XL28 A0A1S4ETD4 A0A182J918 A0A3B3Z9U4 A0A0M3QUP7 A0A1Q3F7Z7 I3KE39 A0A250XW22 U3K8G5 A0A1B6FE18 A0A3B3Z9A2 A0A3P9D6X9 A0A1A9W118 A0A3Q4MXS0 H9GP06 A0A1Q3FIK7 A0A091RTN1 A0A1Q3FI41 A0A1Q3FJC3 A0A091KDT5 A0A3P8Q5B4 A0A3P9D7J4 A0A091LNL9 G1MTN7 A0A1A9XZK9 A0A091TWZ1 D3TRH5 A0A3L8RUI6 V5G7P2 A0A1B0G7E8 H0Z086 A0A091NJ21 A0A3Q4MXQ3 G3U899 E3TGC7 A0A0A0AM39 Q6PBQ5 Q1LWK2 K7FZ56 B4JW85 A0A091MGQ1 B0S701 B4P8F9 A0A093JBD5 A0A3P8PME1 A0A1U8BLH5 Q8BZB2 A0A3B4H8R4 I3MDG5 E1BUI2 A0A091P7J7 V9L0R4 A0A1B6JRG1 A0A2R7WHC0 A0A3Q1CJ39 A0A2Y9QQ67 A0A093EFJ3
EC Number
4.1.1.36
Pubmed
19121390
26354079
22118469
26108605
12364791
14747013
+ More
17210077 20966253 24845553 24438588 24945155 25348373 25315136 18362917 19820115 26369729 24495485 20920257 23761445 17204158 17510324 26227816 22516182 28004739 23537049 25463417 25186727 28087693 21881562 20838655 20353571 30282656 20360741 20634964 23594743 17381049 17994087 17550304 16141072 15489334 21183079 15592404 24402279
17210077 20966253 24845553 24438588 24945155 25348373 25315136 18362917 19820115 26369729 24495485 20920257 23761445 17204158 17510324 26227816 22516182 28004739 23537049 25463417 25186727 28087693 21881562 20838655 20353571 30282656 20360741 20634964 23594743 17381049 17994087 17550304 16141072 15489334 21183079 15592404 24402279
EMBL
BABH01026591
KQ461192
KPJ07024.1
ODYU01010573
SOQ55778.1
KQ459595
+ More
KPI95729.1 NWSH01001682 PCG70431.1 AGBW02009174 OWR51472.1 NEVH01012097 PNF30376.1 GGFK01011864 MBW45185.1 JRES01000049 KNC34561.1 AAAB01008986 EAA00226.3 GGFM01005500 MBW26251.1 APCN01002437 GGFM01007950 MBW28701.1 GANO01003307 JAB56564.1 GGFJ01009562 MBW58703.1 KK853297 KDR08781.1 GGFM01005517 MBW26268.1 ATLV01020327 KE525298 KFB45086.1 GAPW01004702 JAC08896.1 GAKP01001078 JAC57874.1 DS497670 EFA13040.1 GDHF01022732 JAI29582.1 GDAI01002711 JAI14892.1 AXCM01001643 GAMC01008373 JAB98182.1 ADMH02000674 ETN65373.1 DQ440495 CH477658 ABF18528.1 EAT37680.1 EAT37682.1 JTDY01002456 KOB71374.1 BT127116 AEE62078.1 GEZM01074077 JAV64681.1 KB630779 KB631357 ERL83860.1 ERL84259.1 AXCN02001803 DS234212 EDS33257.1 CP012524 ALC41039.1 GFDL01011344 JAV23701.1 AERX01005343 GFFV01004389 JAV35556.1 AGTO01001423 GECZ01021310 JAS48459.1 AAWZ02029030 GFDL01007732 JAV27313.1 KK931024 KFQ45116.1 GFDL01007804 JAV27241.1 GFDL01007443 JAV27602.1 KK739494 KFP38267.1 KK504214 KFP60766.1 KK467948 KFQ82954.1 EZ424027 ADD20303.1 QUSF01000201 RLV87459.1 GALX01002311 JAB66155.1 CCAG010021563 ABQF01049836 KL384642 KFP89486.1 GU589413 ADO29363.1 KL872122 KGL94648.1 BX571883 BC059622 AAH59622.1 BX548060 AGCU01107710 AGCU01107711 AGCU01107712 CH916375 EDV98223.1 KK827306 KFP73756.1 CAQ13581.1 CM000158 EDW91199.1 KK572421 KFW08294.1 AK036042 BC052928 BC089351 AGTP01003568 AADN05000047 KK668637 KFQ03123.1 JW871888 AFP04406.1 GECU01005915 JAT01792.1 KK854807 PTY18909.1 KK367153 KFV41713.1
KPI95729.1 NWSH01001682 PCG70431.1 AGBW02009174 OWR51472.1 NEVH01012097 PNF30376.1 GGFK01011864 MBW45185.1 JRES01000049 KNC34561.1 AAAB01008986 EAA00226.3 GGFM01005500 MBW26251.1 APCN01002437 GGFM01007950 MBW28701.1 GANO01003307 JAB56564.1 GGFJ01009562 MBW58703.1 KK853297 KDR08781.1 GGFM01005517 MBW26268.1 ATLV01020327 KE525298 KFB45086.1 GAPW01004702 JAC08896.1 GAKP01001078 JAC57874.1 DS497670 EFA13040.1 GDHF01022732 JAI29582.1 GDAI01002711 JAI14892.1 AXCM01001643 GAMC01008373 JAB98182.1 ADMH02000674 ETN65373.1 DQ440495 CH477658 ABF18528.1 EAT37680.1 EAT37682.1 JTDY01002456 KOB71374.1 BT127116 AEE62078.1 GEZM01074077 JAV64681.1 KB630779 KB631357 ERL83860.1 ERL84259.1 AXCN02001803 DS234212 EDS33257.1 CP012524 ALC41039.1 GFDL01011344 JAV23701.1 AERX01005343 GFFV01004389 JAV35556.1 AGTO01001423 GECZ01021310 JAS48459.1 AAWZ02029030 GFDL01007732 JAV27313.1 KK931024 KFQ45116.1 GFDL01007804 JAV27241.1 GFDL01007443 JAV27602.1 KK739494 KFP38267.1 KK504214 KFP60766.1 KK467948 KFQ82954.1 EZ424027 ADD20303.1 QUSF01000201 RLV87459.1 GALX01002311 JAB66155.1 CCAG010021563 ABQF01049836 KL384642 KFP89486.1 GU589413 ADO29363.1 KL872122 KGL94648.1 BX571883 BC059622 AAH59622.1 BX548060 AGCU01107710 AGCU01107711 AGCU01107712 CH916375 EDV98223.1 KK827306 KFP73756.1 CAQ13581.1 CM000158 EDW91199.1 KK572421 KFW08294.1 AK036042 BC052928 BC089351 AGTP01003568 AADN05000047 KK668637 KFQ03123.1 JW871888 AFP04406.1 GECU01005915 JAT01792.1 KK854807 PTY18909.1 KK367153 KFV41713.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000075881
+ More
UP000235965 UP000037069 UP000007062 UP000076407 UP000069272 UP000075903 UP000075840 UP000095300 UP000075882 UP000027135 UP000075920 UP000030765 UP000095301 UP000007266 UP000075883 UP000000673 UP000008820 UP000075884 UP000075900 UP000037510 UP000192223 UP000030742 UP000075886 UP000002320 UP000079169 UP000075880 UP000261520 UP000092553 UP000005207 UP000016665 UP000265160 UP000091820 UP000261580 UP000001646 UP000265100 UP000001645 UP000092443 UP000276834 UP000092444 UP000007754 UP000007646 UP000221080 UP000053858 UP000000437 UP000007267 UP000001070 UP000002282 UP000189706 UP000000589 UP000261460 UP000005215 UP000000539 UP000257160 UP000248480
UP000235965 UP000037069 UP000007062 UP000076407 UP000069272 UP000075903 UP000075840 UP000095300 UP000075882 UP000027135 UP000075920 UP000030765 UP000095301 UP000007266 UP000075883 UP000000673 UP000008820 UP000075884 UP000075900 UP000037510 UP000192223 UP000030742 UP000075886 UP000002320 UP000079169 UP000075880 UP000261520 UP000092553 UP000005207 UP000016665 UP000265160 UP000091820 UP000261580 UP000001646 UP000265100 UP000001645 UP000092443 UP000276834 UP000092444 UP000007754 UP000007646 UP000221080 UP000053858 UP000000437 UP000007267 UP000001070 UP000002282 UP000189706 UP000000589 UP000261460 UP000005215 UP000000539 UP000257160 UP000248480
SUPFAM
SSF52507
SSF52507
Gene 3D
ProteinModelPortal
H9JK08
A0A0N0PAJ4
A0A2H1WRW2
A0A194PSJ4
A0A2A4JEQ2
A0A212FCL2
+ More
A0A182KBI3 A0A2J7QP62 A0A2M4AWQ4 A0A0L0CQP0 Q7PZN2 A0A182XFQ3 A0A182FVS7 A0A2M3ZCL9 A0A182UQE4 A0A182HHS1 A0A2M3ZJK1 U5ESC8 A0A2M4C078 A0A1I8NPL5 A0A182KLL6 A0A067QIV1 A0A182W255 A0A2M3ZCS2 A0A084W4E2 A0A023EIV9 A0A034WTA2 A0A1I8MJB3 D2CFX2 A0A0K8USB0 A0A0K8TKK5 A0A182MV92 W8BHG4 W5JPY6 Q1HQE9 A0A182N9P8 A0A182RMQ5 A0A0L7L7Q6 J3JV50 A0A1Y1KW16 A0A1W4WXV5 U4TS01 A0A182R154 B0XL28 A0A1S4ETD4 A0A182J918 A0A3B3Z9U4 A0A0M3QUP7 A0A1Q3F7Z7 I3KE39 A0A250XW22 U3K8G5 A0A1B6FE18 A0A3B3Z9A2 A0A3P9D6X9 A0A1A9W118 A0A3Q4MXS0 H9GP06 A0A1Q3FIK7 A0A091RTN1 A0A1Q3FI41 A0A1Q3FJC3 A0A091KDT5 A0A3P8Q5B4 A0A3P9D7J4 A0A091LNL9 G1MTN7 A0A1A9XZK9 A0A091TWZ1 D3TRH5 A0A3L8RUI6 V5G7P2 A0A1B0G7E8 H0Z086 A0A091NJ21 A0A3Q4MXQ3 G3U899 E3TGC7 A0A0A0AM39 Q6PBQ5 Q1LWK2 K7FZ56 B4JW85 A0A091MGQ1 B0S701 B4P8F9 A0A093JBD5 A0A3P8PME1 A0A1U8BLH5 Q8BZB2 A0A3B4H8R4 I3MDG5 E1BUI2 A0A091P7J7 V9L0R4 A0A1B6JRG1 A0A2R7WHC0 A0A3Q1CJ39 A0A2Y9QQ67 A0A093EFJ3
A0A182KBI3 A0A2J7QP62 A0A2M4AWQ4 A0A0L0CQP0 Q7PZN2 A0A182XFQ3 A0A182FVS7 A0A2M3ZCL9 A0A182UQE4 A0A182HHS1 A0A2M3ZJK1 U5ESC8 A0A2M4C078 A0A1I8NPL5 A0A182KLL6 A0A067QIV1 A0A182W255 A0A2M3ZCS2 A0A084W4E2 A0A023EIV9 A0A034WTA2 A0A1I8MJB3 D2CFX2 A0A0K8USB0 A0A0K8TKK5 A0A182MV92 W8BHG4 W5JPY6 Q1HQE9 A0A182N9P8 A0A182RMQ5 A0A0L7L7Q6 J3JV50 A0A1Y1KW16 A0A1W4WXV5 U4TS01 A0A182R154 B0XL28 A0A1S4ETD4 A0A182J918 A0A3B3Z9U4 A0A0M3QUP7 A0A1Q3F7Z7 I3KE39 A0A250XW22 U3K8G5 A0A1B6FE18 A0A3B3Z9A2 A0A3P9D6X9 A0A1A9W118 A0A3Q4MXS0 H9GP06 A0A1Q3FIK7 A0A091RTN1 A0A1Q3FI41 A0A1Q3FJC3 A0A091KDT5 A0A3P8Q5B4 A0A3P9D7J4 A0A091LNL9 G1MTN7 A0A1A9XZK9 A0A091TWZ1 D3TRH5 A0A3L8RUI6 V5G7P2 A0A1B0G7E8 H0Z086 A0A091NJ21 A0A3Q4MXQ3 G3U899 E3TGC7 A0A0A0AM39 Q6PBQ5 Q1LWK2 K7FZ56 B4JW85 A0A091MGQ1 B0S701 B4P8F9 A0A093JBD5 A0A3P8PME1 A0A1U8BLH5 Q8BZB2 A0A3B4H8R4 I3MDG5 E1BUI2 A0A091P7J7 V9L0R4 A0A1B6JRG1 A0A2R7WHC0 A0A3Q1CJ39 A0A2Y9QQ67 A0A093EFJ3
PDB
1QZU
E-value=4.54981e-58,
Score=565
Ontologies
PATHWAY
GO
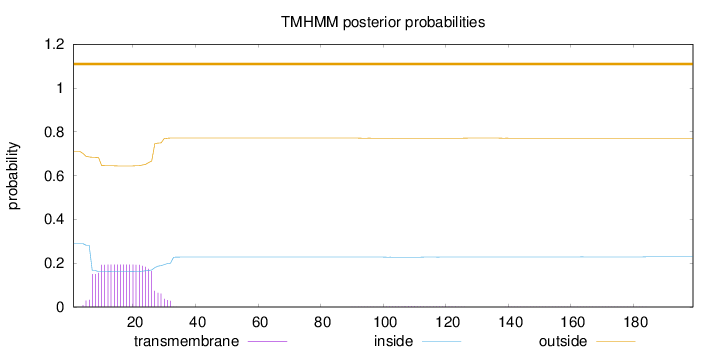
Topology
Length:
199
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.13101
Exp number, first 60 AAs:
4.04809
Total prob of N-in:
0.28927
outside
1 - 199
Population Genetic Test Statistics
Pi
317.884719
Theta
232.292353
Tajima's D
1.16045
CLR
0
CSRT
0.702064896755162
Interpretation
Uncertain
