Gene
KWMTBOMO04750
Pre Gene Modal
BGIBMGA009881
Annotation
PREDICTED:_formin-like_protein_CG32138_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 2.822
Sequence
CDS
ATGGTCGGTGATTCAACATCAACGCAGGTACTCAGAGATCTGGAGATTTCACTACGAACGAATCACATAGAATGGGTGCGAGAGTTCCTCAACGAACAGAACCAAGGATTGGATGTGCTGATAGACTACCTTAGCTTCAGGTTAAGCATGATGAGGCACGAGCAGAGGATAGCTCTCGCCAGGAGCCAGTCCAGCGATGGAATCAATCAAATAAACACAACAACATCGGAATGCAGCGGACCCACAGAGGCGGGTTCCACTTTGAACTCCACTTGGAGGAGGAGAGCCCGAAGCGCCGACTCGGAGATCGAGGGCGCGGCCAGCCCGGCCGTCGCCAGGCGGAGGACGAGGCACGCGGCCAGACTCAACATGGGGGCTTCCACGGACGACATACACGTCTGCATCATGTGCATGAGGGCCATTATGAATAATAAGTATGGCTTCAATATGGTGATCCAACACAGAGAAGCGATAAACAGCATCGCCCTCTCCCTGGTCCACCACTCGCTGCGGACCAAAGCCTTGGTGCTGGAGCTATTGGCGGCCATCTGCCTAGTGAAGGGCGGTCACCAGATCATACTGTCCGCTTTCGATAACTTCAAGGAGGTTATGGGGGAGCCCAGGAGGTTTCACACGCTCATGGAGTACTTCATGAATTACGATGCGTTTCACATTGAATTCATGGTCGCGTGCATGCAGTTCGTGAACATTATCGTGCACTCAGTCGAGGATATGAATTTCAGAGTGCATCTACAGTATGAGTTTACCGCGCTCAAGCTCGACGACTATCTGGAGAGGCTGAGGCTGTGCGAGAGCGACGACCTACAGGTTCAAATCTCGGCGTATCTAGATAACGTGTTTGATGTCGCCGCCCTCATGGAGGACAGCGAGACCAAAACTGCCGCCCTCGAGAAGGTCAACGAACTGGAGGACGAACTAGGACACGCTCACGAGCGGATGGCGGCGCTGGAGCGGGAGGCCATTGCTAAGCTGGCGACACTGGAGGCCGAGCTGGCGCAGGTGAAGCACGAACGAGACCAGCTCGCCGAAGCCAGGAGGCAGGTTGTCGAAGAGGTGTCCACGTTGAGAAGAGCCCAGCAGGACTCTCGGAACAGACAGTCGATGCTGGAGTCCAAGGTGCAGGAACTCGAGTCGCTCACCAAGTCGCTACCTCGAGGAGCCTCCATGGGTGCTATATCGTCACACGCGATGTGCAACGGCATTAGTAACGGTCACGTGTCCCCACCTCCCCCCCCGCCGGCGCCCTCCCTGCCCAACGGCGACCACAAACCCAGTCCCCCACCGCCGCCCCCACCGGCGCCCGCGCCTCCGCCCGCGGCCCCGTGCCCGCCCCCGCCGCCACCCCCTCCCCTGATAAGTTCCGTTCCGCCACCCCCCGCGCCCCCGGCCCCTGGACTCATGGCCCCGCCGCACCACACCGACGCAATGACCATCAAGAGGAAGGTGGACACTAAATATAAATTGCCTACATTGAACTGGGTCGCACTGAAACCTAATCAGGTCCGCGGCACGATCTTCAACGAGCTGGACGAGGAGCGTCCGCGGCGCCGCATCAACTTCGCCGAGTTCGAGGAGAGGTTCCGGCTGGGCGGCGCCGGGCCCGCGCTGCTCGCCGACACGGACTCGCTCGCCTCCTTCCCCAGCAAGCGCTTCCGGAGGCCGGACACGCTCTCGCTGCTCGAACACACCAGGCTCCGGAATATTGCTATTTCCAGAAGGAAACTCGACATCCCAGTCGAGAGAGTAATATCAGCTGTCAACAACCTCGATTTGAAACAACTCCCGCTGGAGAGCGTGGAGATATTGCAGCGCATGGTCCCCACCGAGGCCGAACAGAAGGCGTACAAGGAATACGTGGCCGAGAAGAAGAACATCAACTTGCTGACCGAGGAAGACAAGTTCTTGATGCAGCTGGCTAAGGTCGAGCGCATATCCGCGAAGCTCTCGATAATGAGCTACATGGGCAACTTCTTCGACAACATCCACCTGATCACGCCCCAGATCCACGCGATCATATCCGGCTCGTCGTCGGTCAAGTCCTCGCAGAAGCTGAGGAACGTTCTAGAAATCATCCTGGCGTTCGGCAACTATTTGAACAGCAGCAAACGCGGCCCCGCCTACGGCTTCAAGCTGCAGTCCCTGGACACTTTGATGGATACGAAATCCACGGACAAGAGGATATCGTTGCTGCACTACATCGTGGCGACCGTGAGGCAGAACTTCCCGGAGCTGATGAACTTCGACACTGAGTTGCTGTACATCGATAAAGCGGCCCAAGTGTCGCTGGAGAACGTGGTGGGGGACGTGTGCGAGCTGGAGCGCGGCATGGAGGCGGTGCGGCGCGAGTGCGAGCGCGCGCACGGGCACGTGCTGCGGGACTTCCTCAACAACGCCGCCGACAAGCTGCGGAGGCTGCGCGCCGAGACCAAGCACGCGCAGGACTCGTTCGCGTCGTGCGTGGAGTACTTCGGCGAGGCGCCCCGGGGCTGCGACGCGAACGCTTTCTTTTCGCTGCTCGTTCGTTTCACGAGGGCGTTCAAGCAAGCCGACGCGGAGAACGAGCAACGCCGACGGCTAGAACAAGCCGCCCAGGCCGACAACGCCGATCCAAACAAATCTAAGCAAGCCCAGAAAAAGCAACAGGATGCTGTAATCAACGAATTAAAAACGAAATCACAAGCCGTCGGCGAAAAGAAGCTGCTTCACCAGGACGAAGTGTACAACGGAGCTTTAGAGGACATATTGCACGGACTCCGGAGCGAACCGTACAGACGAGCTGACGCCGTTCGGCGCTCGCATCGCCGCCGCATCAACTCGCACCGACTCTCTAAAGGACTCGACGAACTGGACGTATGA
Protein
MVGDSTSTQVLRDLEISLRTNHIEWVREFLNEQNQGLDVLIDYLSFRLSMMRHEQRIALARSQSSDGINQINTTTSECSGPTEAGSTLNSTWRRRARSADSEIEGAASPAVARRRTRHAARLNMGASTDDIHVCIMCMRAIMNNKYGFNMVIQHREAINSIALSLVHHSLRTKALVLELLAAICLVKGGHQIILSAFDNFKEVMGEPRRFHTLMEYFMNYDAFHIEFMVACMQFVNIIVHSVEDMNFRVHLQYEFTALKLDDYLERLRLCESDDLQVQISAYLDNVFDVAALMEDSETKTAALEKVNELEDELGHAHERMAALEREAIAKLATLEAELAQVKHERDQLAEARRQVVEEVSTLRRAQQDSRNRQSMLESKVQELESLTKSLPRGASMGAISSHAMCNGISNGHVSPPPPPPAPSLPNGDHKPSPPPPPPPAPAPPPAAPCPPPPPPPPLISSVPPPPAPPAPGLMAPPHHTDAMTIKRKVDTKYKLPTLNWVALKPNQVRGTIFNELDEERPRRRINFAEFEERFRLGGAGPALLADTDSLASFPSKRFRRPDTLSLLEHTRLRNIAISRRKLDIPVERVISAVNNLDLKQLPLESVEILQRMVPTEAEQKAYKEYVAEKKNINLLTEEDKFLMQLAKVERISAKLSIMSYMGNFFDNIHLITPQIHAIISGSSSVKSSQKLRNVLEIILAFGNYLNSSKRGPAYGFKLQSLDTLMDTKSTDKRISLLHYIVATVRQNFPELMNFDTELLYIDKAAQVSLENVVGDVCELERGMEAVRRECERAHGHVLRDFLNNAADKLRRLRAETKHAQDSFASCVEYFGEAPRGCDANAFFSLLVRFTRAFKQADAENEQRRRLEQAAQADNADPNKSKQAQKKQQDAVINELKTKSQAVGEKKLLHQDEVYNGALEDILHGLRSEPYRRADAVRRSHRRRINSHRLSKGLDELDV
Summary
Uniprot
Pubmed
Proteomes
PRIDE
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
PDB
4EAH
E-value=1.71295e-94,
Score=887
Ontologies
GO
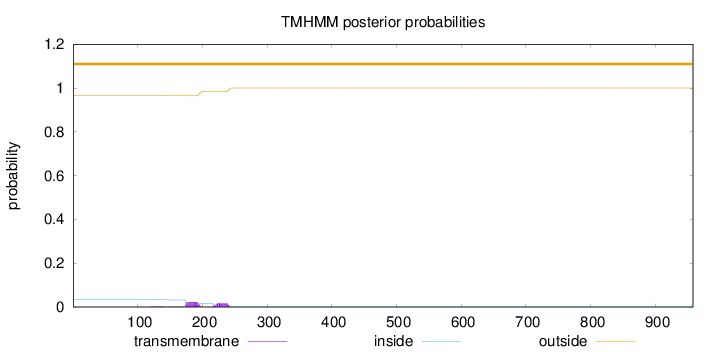
Topology
Length:
958
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.87467
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.03456
outside
1 - 958
Population Genetic Test Statistics
Pi
203.827777
Theta
183.607591
Tajima's D
0.586028
CLR
0.339249
CSRT
0.539273036348183
Interpretation
Uncertain
