Gene
KWMTBOMO04745
Pre Gene Modal
BGIBMGA009879
Annotation
PREDICTED:_CBP80/20-dependent_translation_initiation_factor_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.891
Sequence
CDS
ATGCAGACTCAATTAGGATCAGGCAATGATCTGCAGAGGAGCTACAGGATCCAGGACTTCGCCACGCCTGGCGTCATCATGAGGGAGCGGAGCTTCGTACGTCCTCAGTCGCATCACAACATGCATCATCACCAGTTTTCAAATAATGGGAAACGACGGAGTATGATCCTCACCAACGCAAAGGGAAAACCGACTCTAGAAATCTACAGACCACCAGACGGCGTGGTGTCAACGAACGCTGGTGCCGCCTGTACGACCACCACGGCGGCCGGCAGTGGTGCTGCGGCCACCAACAAGCTGAATGTGCACGCAAAGGAGTTCACAATGGCGAAACCGATTGACCTACACAACAGAGGGGCATTGGGCGTGGGGTACGCTAGTGTGGGACTGCAGCACTCCAGGTCGGCGGTGCTGCACGCGCAGGTGCCGCCCCACTACCGACCCGTGATGCCGCCTCACGCGCTCCTCAATAGCGCCTCCAGCGGGAACGTGCCACATGCACAGGGGCCACGCGTGCACTTCAAGCTGCAACCACAGACTATCATTCCTGAGAAGAAGAAATCCCCTCCGTCGTCGCTGACGAATGGTAACGGGAAGAGCATCCGTCCACAATCGTTTACACCAGGATTGAAAAGGTCAAAGTCCTTAACGTCAGCGGATGCACTCGCAAGTGGCATGGCGGCCCTGGGTCTGGCAGCCGACGCCGGCGACCTTGGTACATTCCCGCCGGAGATTCAAGAGTGTATTGATAAAGCCCTGGAAGATCCGAATGCTGTGTGCGCCCGTACCCTGATGGACGCTGTGGGGCACCTGGTCTCGCGCGCAGTGGAGTCGCCGCGGTACGCGCTGCCGGCCGCCCGCCGCTGCATCGCCGTGGTTGAGCGCGAGCAGACGGAAACCTTCCTCGAGTCGCTGCTTAACACGTGCCAGCAATGGTACCACGATAGGGACAAGCTGTTGGGCGCGATGATCGGCGGTGGTCGACCTCGGTTGATGGCGTTCCTGTCGTTCTTGCTGGAGATGTACTGCCAGCTGCGGCGCCGGGCCATCCAGCGACGGGGAGCCAGCGCCCAGCCCGGGCACGTGCTGCTCGCGCTCATCTGCAAGTGCTGCGAGGACTGCATCAGGCAGCCGGTGCCTTCTGCTAGTGATACTGAGAATCTGTTCTTCGTACTGACGTACATAGGCCGAGACTTGGAAACTCAGCTACCTGGAGATTTGGAGCGATTACTTACAGCCGTCCGTGACGCTTTCCTCAATACTGCCGCGGCTCCTTCCATCAGGCGTACTTTACTGCAGCTTATAGAGCTTCACGCATCCCGGTGGCAGTTACCAGGATGCGCAGTTCTTTATTACTATCCTTCGTCCAAATAG
Protein
MQTQLGSGNDLQRSYRIQDFATPGVIMRERSFVRPQSHHNMHHHQFSNNGKRRSMILTNAKGKPTLEIYRPPDGVVSTNAGAACTTTTAAGSGAAATNKLNVHAKEFTMAKPIDLHNRGALGVGYASVGLQHSRSAVLHAQVPPHYRPVMPPHALLNSASSGNVPHAQGPRVHFKLQPQTIIPEKKKSPPSSLTNGNGKSIRPQSFTPGLKRSKSLTSADALASGMAALGLAADAGDLGTFPPEIQECIDKALEDPNAVCARTLMDAVGHLVSRAVESPRYALPAARRCIAVVEREQTETFLESLLNTCQQWYHDRDKLLGAMIGGGRPRLMAFLSFLLEMYCQLRRRAIQRRGASAQPGHVLLALICKCCEDCIRQPVPSASDTENLFFVLTYIGRDLETQLPGDLERLLTAVRDAFLNTAAAPSIRRTLLQLIELHASRWQLPGCAVLYYYPSSK
Summary
Uniprot
A0A1E1WEH2
A0A1E1WHH8
A0A2H1WG72
A0A212EZ33
A0A0N1IJU5
A0A2A4IV43
+ More
A0A1E1WSX9 A0A1E1WA79 A0A0L7L2R9 A0A182MPE6 A0A2J7QJG6 A0A2J7QJG2 A0A182GJK9 Q172D9 A0A182GN41 A0A182X648 A0A182VTL1 D6WN17 Q7Q875 A0A182P2T9 A0A1S4GZY4 A0A182KWL6 A0A182IB70 A0A182UTU3 A0A1Q3FSD6 A0A182NB79 A0A1Q3FM96 A0A182QXP5 A0A1W4X3W6 A0A1Q3FMZ3 A0A1Q3FKR5 A0A1Q3FK00 A0A182Y8J6 N6TAR4 W5JG83 A0A084VMP9 A0A182RVS3 A0A2J7QJG8 A0A1J1I0G1 A0A023EV52 A0A1L8DRX8 A0A2M4A4Q4 A0A1L8DRY9 A0A2J7QJF4 A0A1L8DS70 A0A1B0CB59 A0A2M4A5U3 A0A182J9H4 A0A182F2S0 A0A2M3Z4V6 A0A1Q3FS67 A0A1Q3FSC9 A0A1S4JW10 A0A336LW89 B0WXF6 A0A1Q3FMJ8 A0A1Q3FPT3 A0A1Q3FJL4 A0A1Q3FM49 A0A1Q3FMA1 A0A1Q3FKJ1 A0A2M4BL62 A0A2J7QJF2 K7IRZ3 A0A1Y1K5R5 A0A336LZB5 A0A0L7R8L4 B4MZD2 U5EZD3 A0A154PQD3 A0A232F103 A0A2A3E319 V9IC82 V9IA14 A0A067QYA5 A0A026WLP1 E9IG66 A0A088AIX7 A0A151XB67 A0A195DJ80 A0A1B6E3M5 F4WBV8 A0A195D3K6 A0A195BEB4 A0A195FSP8 B4G7W0 E1ZXA9 A0A158NQQ2 Q29ND8 W8B6J9 A0A1I8PD07 A0A1I8MGK7 A0A0Q9X1P0 A0A1B0FGA5 T1PEU5 A0A0A1XEF7
A0A1E1WSX9 A0A1E1WA79 A0A0L7L2R9 A0A182MPE6 A0A2J7QJG6 A0A2J7QJG2 A0A182GJK9 Q172D9 A0A182GN41 A0A182X648 A0A182VTL1 D6WN17 Q7Q875 A0A182P2T9 A0A1S4GZY4 A0A182KWL6 A0A182IB70 A0A182UTU3 A0A1Q3FSD6 A0A182NB79 A0A1Q3FM96 A0A182QXP5 A0A1W4X3W6 A0A1Q3FMZ3 A0A1Q3FKR5 A0A1Q3FK00 A0A182Y8J6 N6TAR4 W5JG83 A0A084VMP9 A0A182RVS3 A0A2J7QJG8 A0A1J1I0G1 A0A023EV52 A0A1L8DRX8 A0A2M4A4Q4 A0A1L8DRY9 A0A2J7QJF4 A0A1L8DS70 A0A1B0CB59 A0A2M4A5U3 A0A182J9H4 A0A182F2S0 A0A2M3Z4V6 A0A1Q3FS67 A0A1Q3FSC9 A0A1S4JW10 A0A336LW89 B0WXF6 A0A1Q3FMJ8 A0A1Q3FPT3 A0A1Q3FJL4 A0A1Q3FM49 A0A1Q3FMA1 A0A1Q3FKJ1 A0A2M4BL62 A0A2J7QJF2 K7IRZ3 A0A1Y1K5R5 A0A336LZB5 A0A0L7R8L4 B4MZD2 U5EZD3 A0A154PQD3 A0A232F103 A0A2A3E319 V9IC82 V9IA14 A0A067QYA5 A0A026WLP1 E9IG66 A0A088AIX7 A0A151XB67 A0A195DJ80 A0A1B6E3M5 F4WBV8 A0A195D3K6 A0A195BEB4 A0A195FSP8 B4G7W0 E1ZXA9 A0A158NQQ2 Q29ND8 W8B6J9 A0A1I8PD07 A0A1I8MGK7 A0A0Q9X1P0 A0A1B0FGA5 T1PEU5 A0A0A1XEF7
Pubmed
EMBL
GDQN01005793
JAT85261.1
GDQN01004609
JAT86445.1
ODYU01008442
SOQ52061.1
+ More
AGBW02011376 OWR46759.1 KQ459784 KPJ19960.1 NWSH01007175 PCG63052.1 GDQN01000905 JAT90149.1 GDQN01007283 JAT83771.1 JTDY01003423 KOB69579.1 AXCM01000486 NEVH01013556 PNF28718.1 PNF28717.1 JXUM01068463 KQ562501 KXJ75783.1 CH477441 EAT40846.1 JXUM01075703 JXUM01075704 KQ562901 KXJ74866.1 KQ971343 EFA04338.1 AAAB01008944 EAA10272.3 APCN01000644 GFDL01004526 JAV30519.1 GFDL01006305 JAV28740.1 AXCN02000635 GFDL01006169 JAV28876.1 GFDL01006951 JAV28094.1 GFDL01007095 JAV27950.1 APGK01037602 KB740948 KB632288 ENN77369.1 ERL91485.1 ADMH02001599 ETN61885.1 ATLV01014616 KE524975 KFB39243.1 PNF28716.1 CVRI01000038 CRK93824.1 GAPW01001239 JAC12359.1 GFDF01004861 JAV09223.1 GGFK01002389 MBW35710.1 GFDF01004862 JAV09222.1 PNF28720.1 GFDF01004860 JAV09224.1 AJWK01004845 GGFK01002779 MBW36100.1 GGFM01002806 MBW23557.1 GFDL01004611 JAV30434.1 GFDL01004693 JAV30352.1 UFQT01000229 SSX22080.1 DS232164 EDS36486.1 GFDL01006244 JAV28801.1 GFDL01005562 JAV29483.1 GFDL01007362 JAV27683.1 GFDL01006356 JAV28689.1 GFDL01006368 JAV28677.1 GFDL01006967 JAV28078.1 GGFJ01004668 MBW53809.1 PNF28721.1 GEZM01094204 JAV55811.1 SSX22079.1 KQ414632 KOC67179.1 CH963913 EDW77405.1 GANO01001002 JAB58869.1 KQ434998 KZC13320.1 NNAY01001316 OXU24385.1 KZ288405 PBC26143.1 JR037733 JR037734 AEY57938.1 AEY57939.1 JR037735 AEY57940.1 KK852824 KDR15456.1 KK107154 EZA56873.1 GL762910 EFZ20482.1 KQ982335 KYQ57579.1 KQ980824 KYN12554.1 GEDC01004772 JAS32526.1 GL888066 EGI68386.1 KQ976885 KYN07483.1 KQ976511 KYM82537.1 KQ981280 KYN43471.1 CH479180 EDW28458.1 GL435030 EFN74199.1 ADTU01023460 CH379060 EAL33404.2 GAMC01012328 GAMC01012327 GAMC01012326 JAB94227.1 KRF98667.1 CCAG010000709 KA647261 AFP61890.1 GBXI01004991 JAD09301.1
AGBW02011376 OWR46759.1 KQ459784 KPJ19960.1 NWSH01007175 PCG63052.1 GDQN01000905 JAT90149.1 GDQN01007283 JAT83771.1 JTDY01003423 KOB69579.1 AXCM01000486 NEVH01013556 PNF28718.1 PNF28717.1 JXUM01068463 KQ562501 KXJ75783.1 CH477441 EAT40846.1 JXUM01075703 JXUM01075704 KQ562901 KXJ74866.1 KQ971343 EFA04338.1 AAAB01008944 EAA10272.3 APCN01000644 GFDL01004526 JAV30519.1 GFDL01006305 JAV28740.1 AXCN02000635 GFDL01006169 JAV28876.1 GFDL01006951 JAV28094.1 GFDL01007095 JAV27950.1 APGK01037602 KB740948 KB632288 ENN77369.1 ERL91485.1 ADMH02001599 ETN61885.1 ATLV01014616 KE524975 KFB39243.1 PNF28716.1 CVRI01000038 CRK93824.1 GAPW01001239 JAC12359.1 GFDF01004861 JAV09223.1 GGFK01002389 MBW35710.1 GFDF01004862 JAV09222.1 PNF28720.1 GFDF01004860 JAV09224.1 AJWK01004845 GGFK01002779 MBW36100.1 GGFM01002806 MBW23557.1 GFDL01004611 JAV30434.1 GFDL01004693 JAV30352.1 UFQT01000229 SSX22080.1 DS232164 EDS36486.1 GFDL01006244 JAV28801.1 GFDL01005562 JAV29483.1 GFDL01007362 JAV27683.1 GFDL01006356 JAV28689.1 GFDL01006368 JAV28677.1 GFDL01006967 JAV28078.1 GGFJ01004668 MBW53809.1 PNF28721.1 GEZM01094204 JAV55811.1 SSX22079.1 KQ414632 KOC67179.1 CH963913 EDW77405.1 GANO01001002 JAB58869.1 KQ434998 KZC13320.1 NNAY01001316 OXU24385.1 KZ288405 PBC26143.1 JR037733 JR037734 AEY57938.1 AEY57939.1 JR037735 AEY57940.1 KK852824 KDR15456.1 KK107154 EZA56873.1 GL762910 EFZ20482.1 KQ982335 KYQ57579.1 KQ980824 KYN12554.1 GEDC01004772 JAS32526.1 GL888066 EGI68386.1 KQ976885 KYN07483.1 KQ976511 KYM82537.1 KQ981280 KYN43471.1 CH479180 EDW28458.1 GL435030 EFN74199.1 ADTU01023460 CH379060 EAL33404.2 GAMC01012328 GAMC01012327 GAMC01012326 JAB94227.1 KRF98667.1 CCAG010000709 KA647261 AFP61890.1 GBXI01004991 JAD09301.1
Proteomes
UP000007151
UP000053240
UP000218220
UP000037510
UP000075883
UP000235965
+ More
UP000069940 UP000249989 UP000008820 UP000076407 UP000075920 UP000007266 UP000007062 UP000075885 UP000075882 UP000075840 UP000075903 UP000075884 UP000075886 UP000192223 UP000076408 UP000019118 UP000030742 UP000000673 UP000030765 UP000075900 UP000183832 UP000092461 UP000075880 UP000069272 UP000002320 UP000002358 UP000053825 UP000007798 UP000076502 UP000215335 UP000242457 UP000027135 UP000053097 UP000005203 UP000075809 UP000078492 UP000007755 UP000078542 UP000078540 UP000078541 UP000008744 UP000000311 UP000005205 UP000001819 UP000095300 UP000095301 UP000092444
UP000069940 UP000249989 UP000008820 UP000076407 UP000075920 UP000007266 UP000007062 UP000075885 UP000075882 UP000075840 UP000075903 UP000075884 UP000075886 UP000192223 UP000076408 UP000019118 UP000030742 UP000000673 UP000030765 UP000075900 UP000183832 UP000092461 UP000075880 UP000069272 UP000002320 UP000002358 UP000053825 UP000007798 UP000076502 UP000215335 UP000242457 UP000027135 UP000053097 UP000005203 UP000075809 UP000078492 UP000007755 UP000078542 UP000078540 UP000078541 UP000008744 UP000000311 UP000005205 UP000001819 UP000095300 UP000095301 UP000092444
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A1E1WEH2
A0A1E1WHH8
A0A2H1WG72
A0A212EZ33
A0A0N1IJU5
A0A2A4IV43
+ More
A0A1E1WSX9 A0A1E1WA79 A0A0L7L2R9 A0A182MPE6 A0A2J7QJG6 A0A2J7QJG2 A0A182GJK9 Q172D9 A0A182GN41 A0A182X648 A0A182VTL1 D6WN17 Q7Q875 A0A182P2T9 A0A1S4GZY4 A0A182KWL6 A0A182IB70 A0A182UTU3 A0A1Q3FSD6 A0A182NB79 A0A1Q3FM96 A0A182QXP5 A0A1W4X3W6 A0A1Q3FMZ3 A0A1Q3FKR5 A0A1Q3FK00 A0A182Y8J6 N6TAR4 W5JG83 A0A084VMP9 A0A182RVS3 A0A2J7QJG8 A0A1J1I0G1 A0A023EV52 A0A1L8DRX8 A0A2M4A4Q4 A0A1L8DRY9 A0A2J7QJF4 A0A1L8DS70 A0A1B0CB59 A0A2M4A5U3 A0A182J9H4 A0A182F2S0 A0A2M3Z4V6 A0A1Q3FS67 A0A1Q3FSC9 A0A1S4JW10 A0A336LW89 B0WXF6 A0A1Q3FMJ8 A0A1Q3FPT3 A0A1Q3FJL4 A0A1Q3FM49 A0A1Q3FMA1 A0A1Q3FKJ1 A0A2M4BL62 A0A2J7QJF2 K7IRZ3 A0A1Y1K5R5 A0A336LZB5 A0A0L7R8L4 B4MZD2 U5EZD3 A0A154PQD3 A0A232F103 A0A2A3E319 V9IC82 V9IA14 A0A067QYA5 A0A026WLP1 E9IG66 A0A088AIX7 A0A151XB67 A0A195DJ80 A0A1B6E3M5 F4WBV8 A0A195D3K6 A0A195BEB4 A0A195FSP8 B4G7W0 E1ZXA9 A0A158NQQ2 Q29ND8 W8B6J9 A0A1I8PD07 A0A1I8MGK7 A0A0Q9X1P0 A0A1B0FGA5 T1PEU5 A0A0A1XEF7
A0A1E1WSX9 A0A1E1WA79 A0A0L7L2R9 A0A182MPE6 A0A2J7QJG6 A0A2J7QJG2 A0A182GJK9 Q172D9 A0A182GN41 A0A182X648 A0A182VTL1 D6WN17 Q7Q875 A0A182P2T9 A0A1S4GZY4 A0A182KWL6 A0A182IB70 A0A182UTU3 A0A1Q3FSD6 A0A182NB79 A0A1Q3FM96 A0A182QXP5 A0A1W4X3W6 A0A1Q3FMZ3 A0A1Q3FKR5 A0A1Q3FK00 A0A182Y8J6 N6TAR4 W5JG83 A0A084VMP9 A0A182RVS3 A0A2J7QJG8 A0A1J1I0G1 A0A023EV52 A0A1L8DRX8 A0A2M4A4Q4 A0A1L8DRY9 A0A2J7QJF4 A0A1L8DS70 A0A1B0CB59 A0A2M4A5U3 A0A182J9H4 A0A182F2S0 A0A2M3Z4V6 A0A1Q3FS67 A0A1Q3FSC9 A0A1S4JW10 A0A336LW89 B0WXF6 A0A1Q3FMJ8 A0A1Q3FPT3 A0A1Q3FJL4 A0A1Q3FM49 A0A1Q3FMA1 A0A1Q3FKJ1 A0A2M4BL62 A0A2J7QJF2 K7IRZ3 A0A1Y1K5R5 A0A336LZB5 A0A0L7R8L4 B4MZD2 U5EZD3 A0A154PQD3 A0A232F103 A0A2A3E319 V9IC82 V9IA14 A0A067QYA5 A0A026WLP1 E9IG66 A0A088AIX7 A0A151XB67 A0A195DJ80 A0A1B6E3M5 F4WBV8 A0A195D3K6 A0A195BEB4 A0A195FSP8 B4G7W0 E1ZXA9 A0A158NQQ2 Q29ND8 W8B6J9 A0A1I8PD07 A0A1I8MGK7 A0A0Q9X1P0 A0A1B0FGA5 T1PEU5 A0A0A1XEF7
PDB
4JHK
E-value=8.13272e-06,
Score=119
Ontologies
GO
Topology
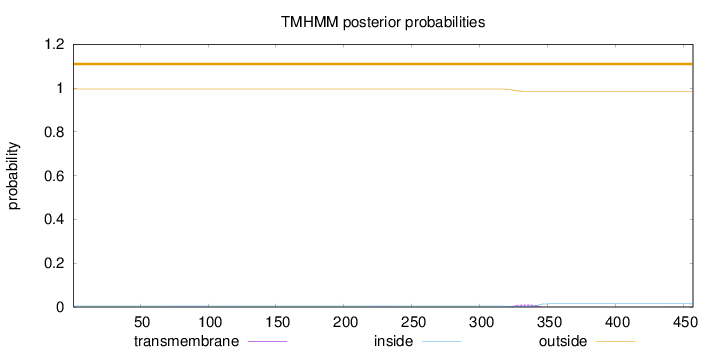
Length:
457
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.23563
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00464
outside
1 - 457
Population Genetic Test Statistics
Pi
294.922095
Theta
179.991439
Tajima's D
1.962321
CLR
0.146914
CSRT
0.87815609219539
Interpretation
Uncertain
