Gene
KWMTBOMO04734
Pre Gene Modal
BGIBMGA009865
Annotation
PREDICTED:_atrial_natriuretic_peptide_receptor_2-like_[Bombyx_mori]
Full name
Guanylate cyclase
Location in the cell
PlasmaMembrane Reliability : 2.479
Sequence
CDS
ATGAAGTCTGCCAGGCGTTCTGTGCTCCTGTACGCGGGTGTTTTAGTCACTGTACTCATCACGTCGGCTGTGCCCTTACTGCTTCTCAGACACGTGGTTTCAACGTTAGAGACATTCACCCACTCATTAAACATGAGTACGGAGAAATTGATAGCCGAGAAACAGAAGAGCGACCTCTTGCTCTCCCGGATGCTCCCGCTGCCAGTTCTGCGGAGGCTGCGGGCACAGAGGACGGTGCCGGCTGAAGCCTTCGACGCGGTCACCATCTTCTTCAGCGATATTGTCGGCTTCACCACCATATCTGCGAATAGCACGCCCATGGAAGTCATCAACATGCTGAACATGCTCTATAAATTGTTTGACGAGAAAATATCTCAATACAACGTTTACAAGGTGGAAACTATAGGAGACGCGTATATGGTGGTCTCTGGTCTGCCACAGAAGAACGGCAGCCGTCACGCGTCCGAGATAGCTGACATGTCATTGTCACTGGTGAGCAGCGTGGAGGAGGCGCGGGTACCGCACCGTCCCGCCGTGCCTCTACGAGTGCGGGCCGGTGTCAACACGGGCCCGTGCGTCGCCGGCGTGGTCGGGACCACCATGCCGAGATACTGCCTTTTTGGCGACACCATCAACACCGCCAGCAGAATGGAGTCTACTGGCGAACCTATGAAAATTCACATCTCGGATTCGACGAAGAGAGCCTTAGACGAGTTCGGTACCTATATCACTGAGTCCAGAGGAATTATTGATATTCCGGGCAAAGGCAGCATGGAAACGTTCTGGTTGCTCGACAAGATCGGAGGAGTACAAAGTGAGAGTCCGCGGTGTCAATTCAACGACTACGACCACAATTTACTGGAACTCATAATTAAATCTTAA
Protein
MKSARRSVLLYAGVLVTVLITSAVPLLLLRHVVSTLETFTHSLNMSTEKLIAEKQKSDLLLSRMLPLPVLRRLRAQRTVPAEAFDAVTIFFSDIVGFTTISANSTPMEVINMLNMLYKLFDEKISQYNVYKVETIGDAYMVVSGLPQKNGSRHASEIADMSLSLVSSVEEARVPHRPAVPLRVRAGVNTGPCVAGVVGTTMPRYCLFGDTINTASRMESTGEPMKIHISDSTKRALDEFGTYITESRGIIDIPGKGSMETFWLLDKIGGVQSESPRCQFNDYDHNLLELIIKS
Summary
Catalytic Activity
GTP = 3',5'-cyclic GMP + diphosphate
Similarity
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Uniprot
H9JK14
A0A194PSH9
A0A0N1PI28
A0A212EVU8
A0A2H1WTN6
A0A0L7LFI9
+ More
A0A1D2NJN9 A0A0P5C8P8 A0A182SK07 A0A0P5N0L5 A0A0P6I899 A0A1S4ELA7 A0A0P6BET9 Q7Q9X3 A0A182TVY7 A0A182MSI9 A0A084WTT8 A0A2H1VQ90 A0A182V862 Q17BS6 A0A182HYT8 A0A182X2J4 A0A182Y8M4 A0A182QBM8 A0A182KZE6 A0A182JVW7 A0A212FDS7 W5JDP6 A0A182W3L0 A0A182PQS0 A0A067R086 A0A182N9U7 A0A2R7WVR9 A0A0N1ICN5 A0A182RDZ4 A0A182F900 A0A164RD13 T1HM71 A0A2J7PHM6 A0A1D2ML76 E9GKU2 A0A2A4JGA7 B0WDM1 A0A1B6HPF3 Q17BS5 A0A1B6MQA4 A0A091HSK3 B0XGU2 A0A1Y3BRU1 Q7Q9X4 G1N3D1 U4TTC7 U3IT17 A0A067R2X5 F4X106 A0A158NI27 A0A0K2TJP5 B4LNM4 A0A0C9QND4 A0A093FJC9 T1GUA8 A0A091RZS2 A0A061QLJ7 A0A091MVY1 L5KCF6 A0A139WCS5 H9GJY6 Q8R1P8 A0A0E9X0L5 A0A093Q8U9 A0A093R4F7 A0A087V9F1 H3AE67 A0A2I0M3A9 A0A226EPN3 B4KM65 A0A3L8S379 A0A091H8N2 A0A091JMP6 A0A091QXK6 A0A091VAH8 A0A0Q3XAF4 V9IKY9 A0A2U9C161 S4PX19 A0A094KSS2 E2BEU4 K7FIE2 A0A1D2NFI3 V5G9K2 E0VPI7 A0A2D0QYK5
A0A1D2NJN9 A0A0P5C8P8 A0A182SK07 A0A0P5N0L5 A0A0P6I899 A0A1S4ELA7 A0A0P6BET9 Q7Q9X3 A0A182TVY7 A0A182MSI9 A0A084WTT8 A0A2H1VQ90 A0A182V862 Q17BS6 A0A182HYT8 A0A182X2J4 A0A182Y8M4 A0A182QBM8 A0A182KZE6 A0A182JVW7 A0A212FDS7 W5JDP6 A0A182W3L0 A0A182PQS0 A0A067R086 A0A182N9U7 A0A2R7WVR9 A0A0N1ICN5 A0A182RDZ4 A0A182F900 A0A164RD13 T1HM71 A0A2J7PHM6 A0A1D2ML76 E9GKU2 A0A2A4JGA7 B0WDM1 A0A1B6HPF3 Q17BS5 A0A1B6MQA4 A0A091HSK3 B0XGU2 A0A1Y3BRU1 Q7Q9X4 G1N3D1 U4TTC7 U3IT17 A0A067R2X5 F4X106 A0A158NI27 A0A0K2TJP5 B4LNM4 A0A0C9QND4 A0A093FJC9 T1GUA8 A0A091RZS2 A0A061QLJ7 A0A091MVY1 L5KCF6 A0A139WCS5 H9GJY6 Q8R1P8 A0A0E9X0L5 A0A093Q8U9 A0A093R4F7 A0A087V9F1 H3AE67 A0A2I0M3A9 A0A226EPN3 B4KM65 A0A3L8S379 A0A091H8N2 A0A091JMP6 A0A091QXK6 A0A091VAH8 A0A0Q3XAF4 V9IKY9 A0A2U9C161 S4PX19 A0A094KSS2 E2BEU4 K7FIE2 A0A1D2NFI3 V5G9K2 E0VPI7 A0A2D0QYK5
EC Number
4.6.1.2
Pubmed
19121390
26354079
22118469
26227816
27289101
12364791
+ More
14747013 17210077 24438588 17510324 25244985 20966253 20920257 23761445 24845553 21292972 20838655 23537049 23749191 21719571 21347285 17994087 23258410 18362917 19820115 21881562 15489334 25613341 9215903 23371554 30282656 23622113 20798317 17381049 20566863
14747013 17210077 24438588 17510324 25244985 20966253 20920257 23761445 24845553 21292972 20838655 23537049 23749191 21719571 21347285 17994087 23258410 18362917 19820115 21881562 15489334 25613341 9215903 23371554 30282656 23622113 20798317 17381049 20566863
EMBL
BABH01026556
BABH01026557
KQ459595
KPI95714.1
KQ459784
KPJ19950.1
+ More
AGBW02012140 OWR45574.1 ODYU01010978 SOQ56430.1 JTDY01001316 KOB74222.1 LJIJ01000028 ODN05186.1 GDIP01173931 JAJ49471.1 GDIQ01149084 JAL02642.1 GDIQ01008120 JAN86617.1 GDIP01030400 JAM73315.1 AAAB01008898 EAA09176.4 AXCM01000458 ATLV01026914 KE525420 KFB53632.1 ODYU01003514 SOQ42404.1 CH477318 EAT43691.1 APCN01005408 AXCN02001201 AGBW02009004 OWR51884.1 ADMH02001458 ETN62492.1 KK853042 KDR12137.1 KK855717 PTY23639.1 KPJ19949.1 LRGB01002190 KZS08548.1 ACPB03017284 NEVH01025135 PNF15831.1 LJIJ01000914 ODM93786.1 GL732550 EFX79739.1 NWSH01001513 PCG71011.1 DS231898 EDS44630.1 GECU01031150 JAS76556.1 EAT43692.1 GEBQ01001932 JAT38045.1 KL217800 KFO98866.1 DS233064 EDS27829.1 MUJZ01002637 OTF83670.1 EAA09177.5 KB630856 KB631023 ERL83905.1 ERL84012.1 ADON01084214 KDR12136.1 GL888520 EGI59866.1 ADTU01016189 ADTU01016190 HACA01008788 CDW26149.1 CH940648 EDW62204.2 GBYB01005079 JAG74846.1 KK390609 KFV54421.1 CAQQ02031877 CAQQ02031878 CAQQ02031879 KK937289 KFQ47905.1 GBFC01000092 JAC59246.1 KK838640 KFP81628.1 KB030846 ELK09022.1 KQ971362 KYB25768.1 BC023420 AAH23420.1 GBXM01012325 JAH96252.1 KL672239 KFW85011.1 KL438435 KFW93530.1 KL484857 KFO09243.1 AFYH01074404 AFYH01074405 AKCR02000044 PKK24170.1 LNIX01000003 OXA58781.1 CH933808 EDW08729.1 QUSF01000077 RLV95590.1 KL525367 KFO90995.1 KK502299 KFP22284.1 KK804800 KFQ31346.1 KK733826 KFR00129.1 LMAW01000094 KQL60805.1 JR050741 AEY61347.1 CP026253 AWP09399.1 GAIX01005808 JAA86752.1 KL266973 KFZ62278.1 GL447857 EFN85810.1 AGCU01118388 AGCU01118389 LJIJ01000057 ODN04011.1 GALX01001646 JAB66820.1 DS235366 EEB15293.1
AGBW02012140 OWR45574.1 ODYU01010978 SOQ56430.1 JTDY01001316 KOB74222.1 LJIJ01000028 ODN05186.1 GDIP01173931 JAJ49471.1 GDIQ01149084 JAL02642.1 GDIQ01008120 JAN86617.1 GDIP01030400 JAM73315.1 AAAB01008898 EAA09176.4 AXCM01000458 ATLV01026914 KE525420 KFB53632.1 ODYU01003514 SOQ42404.1 CH477318 EAT43691.1 APCN01005408 AXCN02001201 AGBW02009004 OWR51884.1 ADMH02001458 ETN62492.1 KK853042 KDR12137.1 KK855717 PTY23639.1 KPJ19949.1 LRGB01002190 KZS08548.1 ACPB03017284 NEVH01025135 PNF15831.1 LJIJ01000914 ODM93786.1 GL732550 EFX79739.1 NWSH01001513 PCG71011.1 DS231898 EDS44630.1 GECU01031150 JAS76556.1 EAT43692.1 GEBQ01001932 JAT38045.1 KL217800 KFO98866.1 DS233064 EDS27829.1 MUJZ01002637 OTF83670.1 EAA09177.5 KB630856 KB631023 ERL83905.1 ERL84012.1 ADON01084214 KDR12136.1 GL888520 EGI59866.1 ADTU01016189 ADTU01016190 HACA01008788 CDW26149.1 CH940648 EDW62204.2 GBYB01005079 JAG74846.1 KK390609 KFV54421.1 CAQQ02031877 CAQQ02031878 CAQQ02031879 KK937289 KFQ47905.1 GBFC01000092 JAC59246.1 KK838640 KFP81628.1 KB030846 ELK09022.1 KQ971362 KYB25768.1 BC023420 AAH23420.1 GBXM01012325 JAH96252.1 KL672239 KFW85011.1 KL438435 KFW93530.1 KL484857 KFO09243.1 AFYH01074404 AFYH01074405 AKCR02000044 PKK24170.1 LNIX01000003 OXA58781.1 CH933808 EDW08729.1 QUSF01000077 RLV95590.1 KL525367 KFO90995.1 KK502299 KFP22284.1 KK804800 KFQ31346.1 KK733826 KFR00129.1 LMAW01000094 KQL60805.1 JR050741 AEY61347.1 CP026253 AWP09399.1 GAIX01005808 JAA86752.1 KL266973 KFZ62278.1 GL447857 EFN85810.1 AGCU01118388 AGCU01118389 LJIJ01000057 ODN04011.1 GALX01001646 JAB66820.1 DS235366 EEB15293.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000037510
UP000094527
+ More
UP000075901 UP000079169 UP000007062 UP000075902 UP000075883 UP000030765 UP000075903 UP000008820 UP000075840 UP000076407 UP000076408 UP000075886 UP000075882 UP000075881 UP000000673 UP000075920 UP000075885 UP000027135 UP000075884 UP000075900 UP000069272 UP000076858 UP000015103 UP000235965 UP000000305 UP000218220 UP000002320 UP000054308 UP000001645 UP000030742 UP000016666 UP000007755 UP000005205 UP000008792 UP000015102 UP000010552 UP000007266 UP000001646 UP000053258 UP000008672 UP000053872 UP000198287 UP000009192 UP000276834 UP000053119 UP000053605 UP000051836 UP000246464 UP000008237 UP000007267 UP000009046 UP000221080
UP000075901 UP000079169 UP000007062 UP000075902 UP000075883 UP000030765 UP000075903 UP000008820 UP000075840 UP000076407 UP000076408 UP000075886 UP000075882 UP000075881 UP000000673 UP000075920 UP000075885 UP000027135 UP000075884 UP000075900 UP000069272 UP000076858 UP000015103 UP000235965 UP000000305 UP000218220 UP000002320 UP000054308 UP000001645 UP000030742 UP000016666 UP000007755 UP000005205 UP000008792 UP000015102 UP000010552 UP000007266 UP000001646 UP000053258 UP000008672 UP000053872 UP000198287 UP000009192 UP000276834 UP000053119 UP000053605 UP000051836 UP000246464 UP000008237 UP000007267 UP000009046 UP000221080
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JK14
A0A194PSH9
A0A0N1PI28
A0A212EVU8
A0A2H1WTN6
A0A0L7LFI9
+ More
A0A1D2NJN9 A0A0P5C8P8 A0A182SK07 A0A0P5N0L5 A0A0P6I899 A0A1S4ELA7 A0A0P6BET9 Q7Q9X3 A0A182TVY7 A0A182MSI9 A0A084WTT8 A0A2H1VQ90 A0A182V862 Q17BS6 A0A182HYT8 A0A182X2J4 A0A182Y8M4 A0A182QBM8 A0A182KZE6 A0A182JVW7 A0A212FDS7 W5JDP6 A0A182W3L0 A0A182PQS0 A0A067R086 A0A182N9U7 A0A2R7WVR9 A0A0N1ICN5 A0A182RDZ4 A0A182F900 A0A164RD13 T1HM71 A0A2J7PHM6 A0A1D2ML76 E9GKU2 A0A2A4JGA7 B0WDM1 A0A1B6HPF3 Q17BS5 A0A1B6MQA4 A0A091HSK3 B0XGU2 A0A1Y3BRU1 Q7Q9X4 G1N3D1 U4TTC7 U3IT17 A0A067R2X5 F4X106 A0A158NI27 A0A0K2TJP5 B4LNM4 A0A0C9QND4 A0A093FJC9 T1GUA8 A0A091RZS2 A0A061QLJ7 A0A091MVY1 L5KCF6 A0A139WCS5 H9GJY6 Q8R1P8 A0A0E9X0L5 A0A093Q8U9 A0A093R4F7 A0A087V9F1 H3AE67 A0A2I0M3A9 A0A226EPN3 B4KM65 A0A3L8S379 A0A091H8N2 A0A091JMP6 A0A091QXK6 A0A091VAH8 A0A0Q3XAF4 V9IKY9 A0A2U9C161 S4PX19 A0A094KSS2 E2BEU4 K7FIE2 A0A1D2NFI3 V5G9K2 E0VPI7 A0A2D0QYK5
A0A1D2NJN9 A0A0P5C8P8 A0A182SK07 A0A0P5N0L5 A0A0P6I899 A0A1S4ELA7 A0A0P6BET9 Q7Q9X3 A0A182TVY7 A0A182MSI9 A0A084WTT8 A0A2H1VQ90 A0A182V862 Q17BS6 A0A182HYT8 A0A182X2J4 A0A182Y8M4 A0A182QBM8 A0A182KZE6 A0A182JVW7 A0A212FDS7 W5JDP6 A0A182W3L0 A0A182PQS0 A0A067R086 A0A182N9U7 A0A2R7WVR9 A0A0N1ICN5 A0A182RDZ4 A0A182F900 A0A164RD13 T1HM71 A0A2J7PHM6 A0A1D2ML76 E9GKU2 A0A2A4JGA7 B0WDM1 A0A1B6HPF3 Q17BS5 A0A1B6MQA4 A0A091HSK3 B0XGU2 A0A1Y3BRU1 Q7Q9X4 G1N3D1 U4TTC7 U3IT17 A0A067R2X5 F4X106 A0A158NI27 A0A0K2TJP5 B4LNM4 A0A0C9QND4 A0A093FJC9 T1GUA8 A0A091RZS2 A0A061QLJ7 A0A091MVY1 L5KCF6 A0A139WCS5 H9GJY6 Q8R1P8 A0A0E9X0L5 A0A093Q8U9 A0A093R4F7 A0A087V9F1 H3AE67 A0A2I0M3A9 A0A226EPN3 B4KM65 A0A3L8S379 A0A091H8N2 A0A091JMP6 A0A091QXK6 A0A091VAH8 A0A0Q3XAF4 V9IKY9 A0A2U9C161 S4PX19 A0A094KSS2 E2BEU4 K7FIE2 A0A1D2NFI3 V5G9K2 E0VPI7 A0A2D0QYK5
PDB
3ET6
E-value=1.10746e-39,
Score=409
Ontologies
GO
Topology
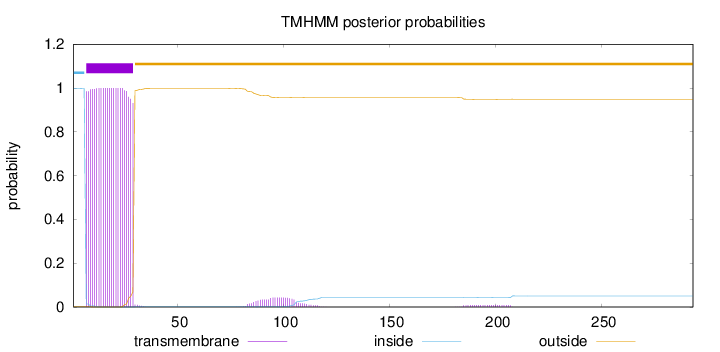
Length:
293
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.95196
Exp number, first 60 AAs:
22.80617
Total prob of N-in:
0.99864
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 293
Population Genetic Test Statistics
Pi
237.296826
Theta
187.428231
Tajima's D
0.68449
CLR
0
CSRT
0.569121543922804
Interpretation
Uncertain
