Gene
KWMTBOMO04733
Pre Gene Modal
BGIBMGA009866
Annotation
PREDICTED:_membrane-bound_alkaline_phosphatase-like_[Bombyx_mori]
Full name
Alkaline phosphatase
+ More
Membrane-bound alkaline phosphatase
Membrane-bound alkaline phosphatase
Location in the cell
Cytoplasmic Reliability : 1.083 Extracellular Reliability : 1.005 Mitochondrial Reliability : 1.519
Sequence
CDS
ATGTTACAGGTTTTAGCTCCTTTAAACAAGAAAGTAGCTAAGAATGTCATCTTATTTCTTGGGGACGGCATGAGCATTACAACGCTCGCCGCCACGAGAGCATACCTCGGCCAGCAGCGGGGACTAAATGGTGAAGAGCTCAAACTGAGTTTCGAAATTTTCCCGTACACCGGCTTAGCTAAGACGTATTGTGTAGATAATAACGTAGCGGACTCTGCGTGTAGTGGTACCGCGTATCTGACTGGAGTGAAGGCCAACAGTGGAACTGTAGGTCTTACCGCCGCCGTCAAGAGAGGAGACTGCGAAGGACAAATTGAAGGCAGACATTCTGTCACCGGACTTATGGATTGGGCGCAACGGGCCGTCAAATCTACTGGTCTCGTGACCACAACACGAGTGACCCACGCCACTCCCGCCGCCGCATACGCTCATTCAGCCGATAGGAGATGGGAAGCTGACACCGATCTTCCAAGCAAAGGCCCTCGCTGCGATGACATAGCGACGCAATTAGTTAGAGGACGTATCGGCAGTAAAATTGATGTGGTCCTTGGTGGAGGTCGACACAATTTCTATCCTAAAAACGAGATAGACCCCGAAGGATACTCCGGTTATCGAAGCGATGGTGTCAATCTAGCAAGGGAATGGTTGATTAAGAAGAAATCGCAGGGACGCACCCCGAAATACCTTTACAATAGAAACGACTTACTGAATCTTAAAGTCAACGAATACGACAGCTTATTAGGTTTGTTCAGCCCTGATCACATGCCGTATCATTTGGAAGCTGACGGTGATGATCCGACGCTGACGGAAATGACTAGGAAAGCCATTGAAATGCTGTCGAAAAACAAAAACGGATTTTTCTTGTTCGTTGAAGGTGGAAGAATAGACACGGCGCATCACGAAACGAAAGCTCGCAAAGCTTTGGATGAGACAGCAGAGTTCGCGAAAGCCGTTGAAGCGGCCGTCAGGATGGTGGACGCCGAAGAGACCCTAATCGTTGTGACATCGGACCACAGTCATACGATGACATACAGTGGATACAGTAAGAGAGGCACCGATATCCTAGGATTTACCAATAAGGTAATTTCTGTTTTATGA
Protein
MLQVLAPLNKKVAKNVILFLGDGMSITTLAATRAYLGQQRGLNGEELKLSFEIFPYTGLAKTYCVDNNVADSACSGTAYLTGVKANSGTVGLTAAVKRGDCEGQIEGRHSVTGLMDWAQRAVKSTGLVTTTRVTHATPAAAYAHSADRRWEADTDLPSKGPRCDDIATQLVRGRIGSKIDVVLGGGRHNFYPKNEIDPEGYSGYRSDGVNLAREWLIKKKSQGRTPKYLYNRNDLLNLKVNEYDSLLGLFSPDHMPYHLEADGDDPTLTEMTRKAIEMLSKNKNGFFLFVEGGRIDTAHHETKARKALDETAEFAKAVEAAVRMVDAEETLIVVTSDHSHTMTYSGYSKRGTDILGFTNKVISVL
Summary
Catalytic Activity
a phosphate monoester + H2O = an alcohol + phosphate
Cofactor
Mg(2+)
Zn(2+)
Zn(2+)
Similarity
Belongs to the alkaline phosphatase family.
Keywords
Cell membrane
Complete proteome
Direct protein sequencing
Glycoprotein
GPI-anchor
Hydrolase
Lipoprotein
Magnesium
Membrane
Metal-binding
Phosphoprotein
Reference proteome
Signal
Zinc
Feature
chain Alkaline phosphatase
propeptide Removed in mature form
propeptide Removed in mature form
Uniprot
H9JK15
A0A2A4JNE4
A0A2H1W0P4
A0A212ESB5
A0A194PQX4
A0A2H5CLH1
+ More
A0A3S2P0P5 A0A0N1IIK6 U5EGZ8 B0W3S9 A0A1Q3EV69 A0A1Q3EV65 A0A0T6BBC6 A0A1B6DLR4 A0A1B0CIG9 A0A182QLW3 A0A3G1LC34 A0A2J7PHL4 A0A182XYI9 A0A182RTZ7 A0A067QRJ2 A0A182JL64 W5JSA4 Q16WV8 A0A182MT69 A0A182P207 A0A182JP07 A0A084VQQ6 A0A1S4FLA4 E0VEB3 A0A182NKM7 A0A182WJL0 A0A182FG53 A0A0P6JST2 A0A182SLH6 A0A182KQV2 A0A182VAE3 A0A182WZ34 A0A182I2X2 Q7QJ58 A0A1B6MN90 A0A1B6MFZ1 A0A1J1ID93 A0A182THI0 A0A2L1IQA0 C0JBW2 A0A2J7PHK8 A0A182GRD2 B2ZZW3 A5A746 H9JH21 A0A1B6DQA4 B2ZZW7 B2ZZX0 P29523 B2ZZX1 C6L8N8 A0A139WCZ9 A0A3S2PC92 A0A2P8YNZ8 A0A1L8DJJ7 A0A2H5CLJ8 A0A2H5CLG8 H9JH27 A0A0K8WLU3 G3GBT4 A0A2L1IQA2 C0JBW1 I6PI83 B2ZZW5 A0A1B6D5A0 A0A194R254 A0A2W1BI04 A0A2H1WA01 A0A0A1WHU6 A0A182QLC7 H9JXN7 C6L8N9 B2ZZW4 A0A0M4F783 A0A1W4X350 Q7KXT0 B2ZZW8 A0A194R2N2 A0A182FNG2 A0A067QT98 B4JEK3 A0A194Q8I8 J9JZX4 B4LW25 A0A1L8DMR0 W8BIN1 B2ZZX2 A0A182PSM3 A0A182R8V1 A0A1L8DMR2 A0A0L0C934 B4K503 A0A1L8DMY4
A0A3S2P0P5 A0A0N1IIK6 U5EGZ8 B0W3S9 A0A1Q3EV69 A0A1Q3EV65 A0A0T6BBC6 A0A1B6DLR4 A0A1B0CIG9 A0A182QLW3 A0A3G1LC34 A0A2J7PHL4 A0A182XYI9 A0A182RTZ7 A0A067QRJ2 A0A182JL64 W5JSA4 Q16WV8 A0A182MT69 A0A182P207 A0A182JP07 A0A084VQQ6 A0A1S4FLA4 E0VEB3 A0A182NKM7 A0A182WJL0 A0A182FG53 A0A0P6JST2 A0A182SLH6 A0A182KQV2 A0A182VAE3 A0A182WZ34 A0A182I2X2 Q7QJ58 A0A1B6MN90 A0A1B6MFZ1 A0A1J1ID93 A0A182THI0 A0A2L1IQA0 C0JBW2 A0A2J7PHK8 A0A182GRD2 B2ZZW3 A5A746 H9JH21 A0A1B6DQA4 B2ZZW7 B2ZZX0 P29523 B2ZZX1 C6L8N8 A0A139WCZ9 A0A3S2PC92 A0A2P8YNZ8 A0A1L8DJJ7 A0A2H5CLJ8 A0A2H5CLG8 H9JH27 A0A0K8WLU3 G3GBT4 A0A2L1IQA2 C0JBW1 I6PI83 B2ZZW5 A0A1B6D5A0 A0A194R254 A0A2W1BI04 A0A2H1WA01 A0A0A1WHU6 A0A182QLC7 H9JXN7 C6L8N9 B2ZZW4 A0A0M4F783 A0A1W4X350 Q7KXT0 B2ZZW8 A0A194R2N2 A0A182FNG2 A0A067QT98 B4JEK3 A0A194Q8I8 J9JZX4 B4LW25 A0A1L8DMR0 W8BIN1 B2ZZX2 A0A182PSM3 A0A182R8V1 A0A1L8DMR2 A0A0L0C934 B4K503 A0A1L8DMY4
EC Number
3.1.3.1
Pubmed
EMBL
BABH01026553
NWSH01000914
PCG73557.1
ODYU01005624
SOQ46661.1
AGBW02012833
+ More
OWR44383.1 KQ459595 KPI95712.1 KY912923 AUH25164.1 RSAL01000066 RVE49395.1 KQ459784 KPJ19948.1 GANO01003229 JAB56642.1 DS231833 EDS32226.1 GFDL01015845 JAV19200.1 GFDL01015846 JAV19199.1 LJIG01002313 KRT84627.1 GEDC01020887 GEDC01010675 GEDC01009792 JAS16411.1 JAS26623.1 JAS27506.1 AJWK01013324 AXCN02000551 KY922835 AUF74484.1 NEVH01025135 PNF15822.1 KK853042 KDR12134.1 ADMH02000578 ETN65800.1 CH477553 EAT39089.1 AXCM01001173 ATLV01015292 KE525006 KFB40300.1 DS235088 EEB11719.1 GDUN01000056 JAN95863.1 APCN01000188 AAAB01008807 EAA03918.4 GEBQ01002563 JAT37414.1 GEBQ01005162 JAT34815.1 CVRI01000047 CRK98187.1 MF741670 AVD96953.1 FJ175381 ACN29683.1 PNF15827.1 JXUM01082264 JXUM01082265 KQ563301 KXJ74098.1 AB379672 BAG41968.1 AB270698 BAF62124.1 BABH01037520 GEDC01009448 JAS27850.1 AB379674 BAG41972.1 AB379676 BAG41975.1 D90454 AB055428 AB379677 BAG41976.1 AB516292 BAH95821.1 KQ971362 KYB25767.1 RSAL01000106 RVE47295.1 PYGN01000459 PSN45953.1 GFDF01007570 JAV06514.1 KY912922 AUH25163.1 KY912924 AUH25165.1 BABH01037533 BABH01037534 GDHF01000489 JAI51825.1 JF825967 AEG79734.1 MF741672 AVD96955.1 FJ175380 ACN29682.1 JF979062 AFI81420.1 AB379673 BAG41970.1 GEDC01016426 JAS20872.1 KQ460870 KPJ11777.1 KZ150047 PZC74488.1 ODYU01007279 SOQ49911.1 GBXI01016061 JAC98230.1 AXCN02000252 BABH01044837 AB516293 BAH95822.1 BAG41969.1 CP012526 ALC47682.1 BAB62746.2 BAG41971.1 BAG41973.1 KPJ11779.1 KDR12135.1 CH916369 EDV93134.1 KQ459562 KPI99720.1 ABLF02034222 ABLF02034224 CH940650 EDW66530.1 GFDF01006336 JAV07748.1 GAMC01005400 JAC01156.1 BAG41977.1 GFDF01006338 JAV07746.1 JRES01000835 KNC27894.1 CH933806 EDW13974.1 GFDF01006337 JAV07747.1
OWR44383.1 KQ459595 KPI95712.1 KY912923 AUH25164.1 RSAL01000066 RVE49395.1 KQ459784 KPJ19948.1 GANO01003229 JAB56642.1 DS231833 EDS32226.1 GFDL01015845 JAV19200.1 GFDL01015846 JAV19199.1 LJIG01002313 KRT84627.1 GEDC01020887 GEDC01010675 GEDC01009792 JAS16411.1 JAS26623.1 JAS27506.1 AJWK01013324 AXCN02000551 KY922835 AUF74484.1 NEVH01025135 PNF15822.1 KK853042 KDR12134.1 ADMH02000578 ETN65800.1 CH477553 EAT39089.1 AXCM01001173 ATLV01015292 KE525006 KFB40300.1 DS235088 EEB11719.1 GDUN01000056 JAN95863.1 APCN01000188 AAAB01008807 EAA03918.4 GEBQ01002563 JAT37414.1 GEBQ01005162 JAT34815.1 CVRI01000047 CRK98187.1 MF741670 AVD96953.1 FJ175381 ACN29683.1 PNF15827.1 JXUM01082264 JXUM01082265 KQ563301 KXJ74098.1 AB379672 BAG41968.1 AB270698 BAF62124.1 BABH01037520 GEDC01009448 JAS27850.1 AB379674 BAG41972.1 AB379676 BAG41975.1 D90454 AB055428 AB379677 BAG41976.1 AB516292 BAH95821.1 KQ971362 KYB25767.1 RSAL01000106 RVE47295.1 PYGN01000459 PSN45953.1 GFDF01007570 JAV06514.1 KY912922 AUH25163.1 KY912924 AUH25165.1 BABH01037533 BABH01037534 GDHF01000489 JAI51825.1 JF825967 AEG79734.1 MF741672 AVD96955.1 FJ175380 ACN29682.1 JF979062 AFI81420.1 AB379673 BAG41970.1 GEDC01016426 JAS20872.1 KQ460870 KPJ11777.1 KZ150047 PZC74488.1 ODYU01007279 SOQ49911.1 GBXI01016061 JAC98230.1 AXCN02000252 BABH01044837 AB516293 BAH95822.1 BAG41969.1 CP012526 ALC47682.1 BAB62746.2 BAG41971.1 BAG41973.1 KPJ11779.1 KDR12135.1 CH916369 EDV93134.1 KQ459562 KPI99720.1 ABLF02034222 ABLF02034224 CH940650 EDW66530.1 GFDF01006336 JAV07748.1 GAMC01005400 JAC01156.1 BAG41977.1 GFDF01006338 JAV07746.1 JRES01000835 KNC27894.1 CH933806 EDW13974.1 GFDF01006337 JAV07747.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000283053
UP000053240
+ More
UP000002320 UP000092461 UP000075886 UP000235965 UP000076408 UP000075900 UP000027135 UP000075880 UP000000673 UP000008820 UP000075883 UP000075885 UP000075881 UP000030765 UP000009046 UP000075884 UP000075920 UP000069272 UP000075901 UP000075882 UP000075903 UP000076407 UP000075840 UP000007062 UP000183832 UP000075902 UP000069940 UP000249989 UP000007266 UP000245037 UP000092553 UP000192223 UP000001070 UP000007819 UP000008792 UP000037069 UP000009192
UP000002320 UP000092461 UP000075886 UP000235965 UP000076408 UP000075900 UP000027135 UP000075880 UP000000673 UP000008820 UP000075883 UP000075885 UP000075881 UP000030765 UP000009046 UP000075884 UP000075920 UP000069272 UP000075901 UP000075882 UP000075903 UP000076407 UP000075840 UP000007062 UP000183832 UP000075902 UP000069940 UP000249989 UP000007266 UP000245037 UP000092553 UP000192223 UP000001070 UP000007819 UP000008792 UP000037069 UP000009192
PRIDE
Pfam
PF00245 Alk_phosphatase
Interpro
SUPFAM
SSF53649
SSF53649
Gene 3D
CDD
ProteinModelPortal
H9JK15
A0A2A4JNE4
A0A2H1W0P4
A0A212ESB5
A0A194PQX4
A0A2H5CLH1
+ More
A0A3S2P0P5 A0A0N1IIK6 U5EGZ8 B0W3S9 A0A1Q3EV69 A0A1Q3EV65 A0A0T6BBC6 A0A1B6DLR4 A0A1B0CIG9 A0A182QLW3 A0A3G1LC34 A0A2J7PHL4 A0A182XYI9 A0A182RTZ7 A0A067QRJ2 A0A182JL64 W5JSA4 Q16WV8 A0A182MT69 A0A182P207 A0A182JP07 A0A084VQQ6 A0A1S4FLA4 E0VEB3 A0A182NKM7 A0A182WJL0 A0A182FG53 A0A0P6JST2 A0A182SLH6 A0A182KQV2 A0A182VAE3 A0A182WZ34 A0A182I2X2 Q7QJ58 A0A1B6MN90 A0A1B6MFZ1 A0A1J1ID93 A0A182THI0 A0A2L1IQA0 C0JBW2 A0A2J7PHK8 A0A182GRD2 B2ZZW3 A5A746 H9JH21 A0A1B6DQA4 B2ZZW7 B2ZZX0 P29523 B2ZZX1 C6L8N8 A0A139WCZ9 A0A3S2PC92 A0A2P8YNZ8 A0A1L8DJJ7 A0A2H5CLJ8 A0A2H5CLG8 H9JH27 A0A0K8WLU3 G3GBT4 A0A2L1IQA2 C0JBW1 I6PI83 B2ZZW5 A0A1B6D5A0 A0A194R254 A0A2W1BI04 A0A2H1WA01 A0A0A1WHU6 A0A182QLC7 H9JXN7 C6L8N9 B2ZZW4 A0A0M4F783 A0A1W4X350 Q7KXT0 B2ZZW8 A0A194R2N2 A0A182FNG2 A0A067QT98 B4JEK3 A0A194Q8I8 J9JZX4 B4LW25 A0A1L8DMR0 W8BIN1 B2ZZX2 A0A182PSM3 A0A182R8V1 A0A1L8DMR2 A0A0L0C934 B4K503 A0A1L8DMY4
A0A3S2P0P5 A0A0N1IIK6 U5EGZ8 B0W3S9 A0A1Q3EV69 A0A1Q3EV65 A0A0T6BBC6 A0A1B6DLR4 A0A1B0CIG9 A0A182QLW3 A0A3G1LC34 A0A2J7PHL4 A0A182XYI9 A0A182RTZ7 A0A067QRJ2 A0A182JL64 W5JSA4 Q16WV8 A0A182MT69 A0A182P207 A0A182JP07 A0A084VQQ6 A0A1S4FLA4 E0VEB3 A0A182NKM7 A0A182WJL0 A0A182FG53 A0A0P6JST2 A0A182SLH6 A0A182KQV2 A0A182VAE3 A0A182WZ34 A0A182I2X2 Q7QJ58 A0A1B6MN90 A0A1B6MFZ1 A0A1J1ID93 A0A182THI0 A0A2L1IQA0 C0JBW2 A0A2J7PHK8 A0A182GRD2 B2ZZW3 A5A746 H9JH21 A0A1B6DQA4 B2ZZW7 B2ZZX0 P29523 B2ZZX1 C6L8N8 A0A139WCZ9 A0A3S2PC92 A0A2P8YNZ8 A0A1L8DJJ7 A0A2H5CLJ8 A0A2H5CLG8 H9JH27 A0A0K8WLU3 G3GBT4 A0A2L1IQA2 C0JBW1 I6PI83 B2ZZW5 A0A1B6D5A0 A0A194R254 A0A2W1BI04 A0A2H1WA01 A0A0A1WHU6 A0A182QLC7 H9JXN7 C6L8N9 B2ZZW4 A0A0M4F783 A0A1W4X350 Q7KXT0 B2ZZW8 A0A194R2N2 A0A182FNG2 A0A067QT98 B4JEK3 A0A194Q8I8 J9JZX4 B4LW25 A0A1L8DMR0 W8BIN1 B2ZZX2 A0A182PSM3 A0A182R8V1 A0A1L8DMR2 A0A0L0C934 B4K503 A0A1L8DMY4
PDB
1SHQ
E-value=2.06659e-83,
Score=787
Ontologies
PATHWAY
GO
PANTHER
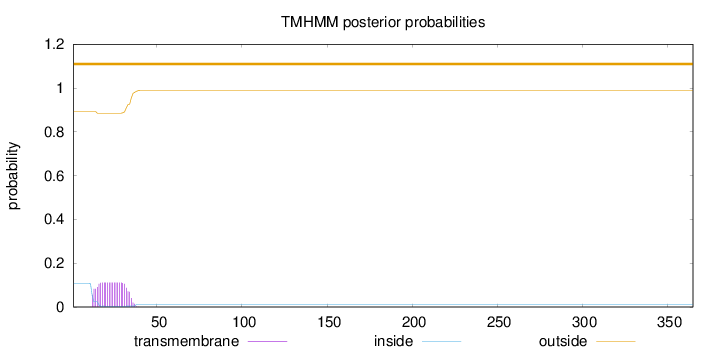
Topology
Subcellular location
Cell membrane
Length:
365
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.38664
Exp number, first 60 AAs:
2.38525
Total prob of N-in:
0.10665
outside
1 - 365
Population Genetic Test Statistics
Pi
142.62143
Theta
126.799601
Tajima's D
-0.517305
CLR
0.497816
CSRT
0.24158792060397
Interpretation
Uncertain
