Gene
KWMTBOMO04727
Pre Gene Modal
BGIBMGA012813
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.012
Sequence
CDS
ATGGATGGAAGGCCAAACGTACGGAAGCTTGCTGAGCCCGACACACGAGCGAAGACCCACACCGCTATTAATCAATGGGCCAACGATCGTCGCACTGAAACCACTGCCTCTGACCTCGACACGTGGACTAGTCTAAAGCGAAGTGTAAAAGATATATGTCACCAATACCTCCAACCATCACAACTGGTCAAAAAACAAGATTGGATGACAGATTCAATTTTGCAGCTTATGGAAGACAGGAGGCAATTTAAGGGTACAAATCCAGTAATGTATAAGCAAATCGATGTCACCATTCGAAAAGAGATTGCGAAAGCCAAGAATGATTACTACAGCAATAAGTGCGCGCACTTGGAAGATCTCCTTCGTAAACATGACAGCTTCAATGCGCACAAGCACATCAAGGAAATGGTTGGTCATAGACGGAAATCAATGAATACTCTGACCGATGCAGGTGGTAACCTCATTCTTGACACCGATGGCAAAAAGAGAGTGTGGTGTGACTATGTTGCAGAGATGTTTGCGGACTCATCGAGGAACATTTCACCGATCACTGATCGGACTGGTCCAGCTATGTTGCGATCTGAGGTTTTGCGGGCCTTGAAAGCTGCTAAAACAGGTAAATCACCGGGTCCTGATAATATACCGATCGAAATACTAAAGTTGCTGGAAGAAGATCAACTGGACGTTCTCACACAGTTCTTCAACAACATCTACGAAACAGGCAATCTTCCTAGTGACTGGCTAAAATCCACATTCATCACGATACCCAAGAAGCAAAATGCCAAAAAATGTGGAGAGTATCGTATGATTAGTCTAATGAGCCATGTCCTAAAAGTATTCTTGAACATCATACAGAATAGAATACGGCCAAAATGTGATGAACAACTGGGCGATAGCCAATTTGGATTTCGATCTGGTGTTGGCACCAGAGAAGCTCTTTTCGCTCTCAACGTTCTCGTACAGAAATGCAGAGACATGCAAACTGATGTCTTTTTGTGCTTCATAGACTACGAAAAGGCATTTGACCGAGTTAAACATCACCAACTTTTTAGCCTTTTATGTGACATTGGTCTAGATGGTAAGGACGTCAGAATCATCCGCAACCTGTACGAGAAGCAGGTGGCTACGATAAGAGTCGAAAACGAGGAAACCGACCAAGTTGAGATCTGTCGTGGGGTCCGGCAGGGGTGTGTCTTATCCCCGTTGCTTTTTAACATCTACTCAGAGGCTGTGATGTCAAAAGCTCTTGAAAATCTTGAAGTGGGAATAGGAATCAACGGCAGGGTCGTTAACAACTTGCGTTATGCGGACGACACTATCCTTATTGCAGCATCGGAAGCCGATTTGCAAGCAATTGTGAATAAGGTTAATGAGTGCAGCGAAGAAGCCGGGTTGTCGATAAACATAAGCAAGACCAAGTTCACGGTAGTCTCAAGAAACCCGGATTTGAGTTCAAGCGTATCTGTAGCTGGGAAACAACTAGAGCGGGTACGACAGTACAAGTATTTAGGGGCTTGGGTAAACGAAGCATGGGAATCTGATCAAGAAATCAAGACCCGGATAGAAACTGCTCGAGCCTCTTTTAATAAAATGAGGAAAGTTCTTTGCTGTCGTCAACTGAATATAAAGCTCCGAGTCAGGCTACTCATGTGCTATATCTGGCCTATCGTCATGTATGGTTGCGAAACGTGGACCCTGAAAGAGGATACTACAAGACGCCTACAGGCATTCGAGATGTGGTGCTACCGTCGCATGTTACGCATTGCAGATGTACTCCTGGCCAAGACGAACCACAGACTGAAAAATATGGCGCAAAATCAGTATATTTTACACGACGCAATGAAAAAATATGGTTAG
Protein
MDGRPNVRKLAEPDTRAKTHTAINQWANDRRTETTASDLDTWTSLKRSVKDICHQYLQPSQLVKKQDWMTDSILQLMEDRRQFKGTNPVMYKQIDVTIRKEIAKAKNDYYSNKCAHLEDLLRKHDSFNAHKHIKEMVGHRRKSMNTLTDAGGNLILDTDGKKRVWCDYVAEMFADSSRNISPITDRTGPAMLRSEVLRALKAAKTGKSPGPDNIPIEILKLLEEDQLDVLTQFFNNIYETGNLPSDWLKSTFITIPKKQNAKKCGEYRMISLMSHVLKVFLNIIQNRIRPKCDEQLGDSQFGFRSGVGTREALFALNVLVQKCRDMQTDVFLCFIDYEKAFDRVKHHQLFSLLCDIGLDGKDVRIIRNLYEKQVATIRVENEETDQVEICRGVRQGCVLSPLLFNIYSEAVMSKALENLEVGIGINGRVVNNLRYADDTILIAASEADLQAIVNKVNECSEEAGLSINISKTKFTVVSRNPDLSSSVSVAGKQLERVRQYKYLGAWVNEAWESDQEIKTRIETARASFNKMRKVLCCRQLNIKLRVRLLMCYIWPIVMYGCETWTLKEDTTRRLQAFEMWCYRRMLRIADVLLAKTNHRLKNMAQNQYILHDAMKKYG
Summary
Uniprot
D5LB38
A0A2A4JBK9
A0A0J7KZE8
A0A2S2QAF2
A0A3S0ZV17
A0A2H6KKM9
+ More
A0A2J7RLC6 A0A2J7PHX9 A0A2J7RNM5 A0A2J7QMH0 A0A2J7PJ38 A0A2J7R3B7 A0A2J7R639 A0A2J7R7D2 A0A2J7QLX7 A0A2J7R696 A0A2J7QQQ3 A0A2J7R4W1 A0A2J7RLG7 A0A2J7RRE4 A0A2J7PJ03 A0A2J7PF68 A0A2J7QNQ3 A0A2J7PZN7 A0A2J7RBL5 A0A2J7PBF1 A0A2J7RBL1 A0A2J7QSZ5 A0A2J7Q649 A0A2J7RPX5 A0A2J7PC89 A0A2J7RAC6 A0A2J7PQH3 A0A2J7R8W8 A0A2J7QJ13 A0A2J7QCG2 A0A2J7QEJ0 A0A2J7RBD8 A0A2J7RBX3 A0A2J7Q2Y3 A0A2J7PY33 A0A2J7Q2T8 A0A2J7PVW0 A0A2J7PT27 A0A2J7R8P5 A0A2J7R901 A0A2J7QUD0 A0A2J7R8G1 A0A2J7QJJ9 A0A2J7QMC5 A0A2J7QI56 A0A2J7Q6P7 A0A2J7RIL8 A0A2J7PNH4 A0A2J7PWV5 A0A3Q1LQN1 A0A2J7REE7 A0A2J7Q3K9 A0A2J7QCG0 A0A2J7QGX8 A0A2J7PTY0 A0A2J7QR01 A0A2J7PW81 A0A2J7PX03 A0A2J7RL19 A0A1W7RAL1 A0A2J7PZJ9 A0A2J7PQA3 A0A2S2PMS6 A0A2J7RLI7 A0A2J7PMJ3 A0A2J7Q2Z3 A0A2J7Q5D5 A0A2J7RP47 A0A2V9X8L4 A0A2J7QGT4 A0A2J7RGX2 A0A2J7Q7C6 A0A2J7Q6A8 A0A2J7Q0C6 A0A2J7QRF9 A0A2J7QH51 A0A2J7QKB0 A0A2J7R4R4 A0A2J7QUN7 A0A3Q1M6J7 A0A2J7Q3T8 A0A2J7QGJ2 A0A2J7PHM9 A0A3Q1LX64 A0A2J7PTS1 A0A2J7R2D1 A0A2J7QSW0 A0A2J7Q647 A0A2J7Q0P9 A0A2J7QYI2 A0A2J7RJ94 A0A2J7RQT0 A0A2J7Q5X4
A0A2J7RLC6 A0A2J7PHX9 A0A2J7RNM5 A0A2J7QMH0 A0A2J7PJ38 A0A2J7R3B7 A0A2J7R639 A0A2J7R7D2 A0A2J7QLX7 A0A2J7R696 A0A2J7QQQ3 A0A2J7R4W1 A0A2J7RLG7 A0A2J7RRE4 A0A2J7PJ03 A0A2J7PF68 A0A2J7QNQ3 A0A2J7PZN7 A0A2J7RBL5 A0A2J7PBF1 A0A2J7RBL1 A0A2J7QSZ5 A0A2J7Q649 A0A2J7RPX5 A0A2J7PC89 A0A2J7RAC6 A0A2J7PQH3 A0A2J7R8W8 A0A2J7QJ13 A0A2J7QCG2 A0A2J7QEJ0 A0A2J7RBD8 A0A2J7RBX3 A0A2J7Q2Y3 A0A2J7PY33 A0A2J7Q2T8 A0A2J7PVW0 A0A2J7PT27 A0A2J7R8P5 A0A2J7R901 A0A2J7QUD0 A0A2J7R8G1 A0A2J7QJJ9 A0A2J7QMC5 A0A2J7QI56 A0A2J7Q6P7 A0A2J7RIL8 A0A2J7PNH4 A0A2J7PWV5 A0A3Q1LQN1 A0A2J7REE7 A0A2J7Q3K9 A0A2J7QCG0 A0A2J7QGX8 A0A2J7PTY0 A0A2J7QR01 A0A2J7PW81 A0A2J7PX03 A0A2J7RL19 A0A1W7RAL1 A0A2J7PZJ9 A0A2J7PQA3 A0A2S2PMS6 A0A2J7RLI7 A0A2J7PMJ3 A0A2J7Q2Z3 A0A2J7Q5D5 A0A2J7RP47 A0A2V9X8L4 A0A2J7QGT4 A0A2J7RGX2 A0A2J7Q7C6 A0A2J7Q6A8 A0A2J7Q0C6 A0A2J7QRF9 A0A2J7QH51 A0A2J7QKB0 A0A2J7R4R4 A0A2J7QUN7 A0A3Q1M6J7 A0A2J7Q3T8 A0A2J7QGJ2 A0A2J7PHM9 A0A3Q1LX64 A0A2J7PTS1 A0A2J7R2D1 A0A2J7QSW0 A0A2J7Q647 A0A2J7Q0P9 A0A2J7QYI2 A0A2J7RJ94 A0A2J7RQT0 A0A2J7Q5X4
EMBL
GU815089
ADF18552.1
NWSH01001998
PCG69477.1
LBMM01001656
KMQ95932.1
+ More
GGMS01004979 MBY74182.1 RQTK01000700 RUS75926.1 BDSA01000081 GBE63537.1 NEVH01002690 PNF41636.1 NEVH01025130 PNF15934.1 NEVH01002150 PNF42436.1 NEVH01013208 PNF29772.1 NEVH01024952 PNF16338.1 NEVH01007822 PNF35313.1 NEVH01006981 PNF36299.1 NEVH01006734 PNF36732.1 NEVH01013241 PNF29590.1 NEVH01006936 PNF36357.1 NEVH01012083 PNF30919.1 NEVH01007402 PNF35873.1 NEVH01002684 PNF41680.1 NEVH01000609 PNF43402.1 PNF16321.1 NEVH01026085 PNF14968.1 NEVH01012564 PNF30212.1 NEVH01020333 PNF21790.1 NEVH01005896 PNF38212.1 NEVH01027104 PNF13657.1 PNF38208.1 NEVH01011205 PNF31711.1 NEVH01017540 PNF24056.1 NEVH01020851 NEVH01013954 NEVH01006574 NEVH01001358 NEVH01001354 PNF21237.1 PNF28332.1 PNF37782.1 PNF42792.1 PNF42884.1 NEVH01027067 PNF13944.1 PNF37787.1 NEVH01022636 PNF18588.1 NEVH01024428 NEVH01017533 NEVH01016946 NEVH01015820 NEVH01015300 NEVH01013960 NEVH01010478 NEVH01010477 NEVH01006721 NEVH01005894 NEVH01003749 NEVH01003737 NEVH01000598 PNF17088.1 PNF24133.1 PNF25000.1 PNF26655.1 PNF27272.1 PNF28247.1 PNF32492.1 PNF32520.1 PNF37274.1 PNF38220.1 PNF40102.1 PNF40402.1 PNF43530.1 NEVH01013563 PNF28562.1 NEVH01016289 PNF26277.1 NEVH01015309 PNF27009.1 NEVH01005906 PNF38146.1 NEVH01005889 PNF38332.1 NEVH01019075 PNF22938.1 NEVH01020850 PNF21258.1 NEVH01019077 PNF22889.1 NEVH01020940 PNF20478.1 NEVH01021921 PNF19488.1 PNF37211.1 PNF37310.1 NEVH01011186 PNF32183.1 NEVH01016335 PNF25462.1 PNF37115.1 NEVH01013553 PNF28765.1 NEVH01013215 PNF29703.1 NEVH01013959 PNF28275.1 NEVH01017460 PNF24248.1 NEVH01003500 PNF40666.1 NEVH01023953 PNF17877.1 NEVH01020867 PNF20816.1 NEVH01005277 PNF39212.1 NEVH01019066 PNF23173.1 PNF26276.1 NEVH01014359 PNF27813.1 NEVH01021219 PNF19787.1 NEVH01012082 PNF31006.1 NEVH01020937 PNF20581.1 NEVH01020865 PNF20863.1 NEVH01002700 PNF41535.1 GFAH01000222 JAV48167.1 PNF21767.1 NEVH01022640 NEVH01008218 NEVH01005885 PNF18513.1 PNF34430.1 PNF38481.1 GGMR01018096 MBY30715.1 PNF41704.1 NEVH01023985 PNF17546.1 PNF22955.1 NEVH01017570 PNF23798.1 NEVH01002141 PNF42599.1 QIAP01000240 PYX61602.1 PNF27789.1 NEVH01003750 PNF40091.1 NEVH01017443 PNF24485.1 NEVH01017534 PNF24114.1 NEVH01019971 PNF22037.1 NEVH01011898 PNF31156.1 NEVH01014358 PNF27914.1 NEVH01013275 PNF29009.1 PNF35815.1 NEVH01023960 NEVH01010501 PNF17732.1 PNF32290.1 NEVH01019064 PNF23230.1 NEVH01018372 NEVH01014371 NEVH01011208 NEVH01002697 NEVH01002552 PNF23678.1 PNF27673.1 PNF31697.1 PNF41592.1 PNF42118.1 NEVH01025135 PNF15828.1 NEVH01021221 PNF19729.1 NEVH01007834 PNF34995.1 NEVH01011217 PNF31657.1 NEVH01017541 NEVH01006570 NEVH01001345 PNF24053.1 PNF37832.1 PNF42982.1 NEVH01019964 PNF22170.1 NEVH01009090 PNF33636.1 NEVH01002994 PNF40909.1 NEVH01000618 PNF43201.1 NEVH01017543 PNF23975.1
GGMS01004979 MBY74182.1 RQTK01000700 RUS75926.1 BDSA01000081 GBE63537.1 NEVH01002690 PNF41636.1 NEVH01025130 PNF15934.1 NEVH01002150 PNF42436.1 NEVH01013208 PNF29772.1 NEVH01024952 PNF16338.1 NEVH01007822 PNF35313.1 NEVH01006981 PNF36299.1 NEVH01006734 PNF36732.1 NEVH01013241 PNF29590.1 NEVH01006936 PNF36357.1 NEVH01012083 PNF30919.1 NEVH01007402 PNF35873.1 NEVH01002684 PNF41680.1 NEVH01000609 PNF43402.1 PNF16321.1 NEVH01026085 PNF14968.1 NEVH01012564 PNF30212.1 NEVH01020333 PNF21790.1 NEVH01005896 PNF38212.1 NEVH01027104 PNF13657.1 PNF38208.1 NEVH01011205 PNF31711.1 NEVH01017540 PNF24056.1 NEVH01020851 NEVH01013954 NEVH01006574 NEVH01001358 NEVH01001354 PNF21237.1 PNF28332.1 PNF37782.1 PNF42792.1 PNF42884.1 NEVH01027067 PNF13944.1 PNF37787.1 NEVH01022636 PNF18588.1 NEVH01024428 NEVH01017533 NEVH01016946 NEVH01015820 NEVH01015300 NEVH01013960 NEVH01010478 NEVH01010477 NEVH01006721 NEVH01005894 NEVH01003749 NEVH01003737 NEVH01000598 PNF17088.1 PNF24133.1 PNF25000.1 PNF26655.1 PNF27272.1 PNF28247.1 PNF32492.1 PNF32520.1 PNF37274.1 PNF38220.1 PNF40102.1 PNF40402.1 PNF43530.1 NEVH01013563 PNF28562.1 NEVH01016289 PNF26277.1 NEVH01015309 PNF27009.1 NEVH01005906 PNF38146.1 NEVH01005889 PNF38332.1 NEVH01019075 PNF22938.1 NEVH01020850 PNF21258.1 NEVH01019077 PNF22889.1 NEVH01020940 PNF20478.1 NEVH01021921 PNF19488.1 PNF37211.1 PNF37310.1 NEVH01011186 PNF32183.1 NEVH01016335 PNF25462.1 PNF37115.1 NEVH01013553 PNF28765.1 NEVH01013215 PNF29703.1 NEVH01013959 PNF28275.1 NEVH01017460 PNF24248.1 NEVH01003500 PNF40666.1 NEVH01023953 PNF17877.1 NEVH01020867 PNF20816.1 NEVH01005277 PNF39212.1 NEVH01019066 PNF23173.1 PNF26276.1 NEVH01014359 PNF27813.1 NEVH01021219 PNF19787.1 NEVH01012082 PNF31006.1 NEVH01020937 PNF20581.1 NEVH01020865 PNF20863.1 NEVH01002700 PNF41535.1 GFAH01000222 JAV48167.1 PNF21767.1 NEVH01022640 NEVH01008218 NEVH01005885 PNF18513.1 PNF34430.1 PNF38481.1 GGMR01018096 MBY30715.1 PNF41704.1 NEVH01023985 PNF17546.1 PNF22955.1 NEVH01017570 PNF23798.1 NEVH01002141 PNF42599.1 QIAP01000240 PYX61602.1 PNF27789.1 NEVH01003750 PNF40091.1 NEVH01017443 PNF24485.1 NEVH01017534 PNF24114.1 NEVH01019971 PNF22037.1 NEVH01011898 PNF31156.1 NEVH01014358 PNF27914.1 NEVH01013275 PNF29009.1 PNF35815.1 NEVH01023960 NEVH01010501 PNF17732.1 PNF32290.1 NEVH01019064 PNF23230.1 NEVH01018372 NEVH01014371 NEVH01011208 NEVH01002697 NEVH01002552 PNF23678.1 PNF27673.1 PNF31697.1 PNF41592.1 PNF42118.1 NEVH01025135 PNF15828.1 NEVH01021221 PNF19729.1 NEVH01007834 PNF34995.1 NEVH01011217 PNF31657.1 NEVH01017541 NEVH01006570 NEVH01001345 PNF24053.1 PNF37832.1 PNF42982.1 NEVH01019964 PNF22170.1 NEVH01009090 PNF33636.1 NEVH01002994 PNF40909.1 NEVH01000618 PNF43201.1 NEVH01017543 PNF23975.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
D5LB38
A0A2A4JBK9
A0A0J7KZE8
A0A2S2QAF2
A0A3S0ZV17
A0A2H6KKM9
+ More
A0A2J7RLC6 A0A2J7PHX9 A0A2J7RNM5 A0A2J7QMH0 A0A2J7PJ38 A0A2J7R3B7 A0A2J7R639 A0A2J7R7D2 A0A2J7QLX7 A0A2J7R696 A0A2J7QQQ3 A0A2J7R4W1 A0A2J7RLG7 A0A2J7RRE4 A0A2J7PJ03 A0A2J7PF68 A0A2J7QNQ3 A0A2J7PZN7 A0A2J7RBL5 A0A2J7PBF1 A0A2J7RBL1 A0A2J7QSZ5 A0A2J7Q649 A0A2J7RPX5 A0A2J7PC89 A0A2J7RAC6 A0A2J7PQH3 A0A2J7R8W8 A0A2J7QJ13 A0A2J7QCG2 A0A2J7QEJ0 A0A2J7RBD8 A0A2J7RBX3 A0A2J7Q2Y3 A0A2J7PY33 A0A2J7Q2T8 A0A2J7PVW0 A0A2J7PT27 A0A2J7R8P5 A0A2J7R901 A0A2J7QUD0 A0A2J7R8G1 A0A2J7QJJ9 A0A2J7QMC5 A0A2J7QI56 A0A2J7Q6P7 A0A2J7RIL8 A0A2J7PNH4 A0A2J7PWV5 A0A3Q1LQN1 A0A2J7REE7 A0A2J7Q3K9 A0A2J7QCG0 A0A2J7QGX8 A0A2J7PTY0 A0A2J7QR01 A0A2J7PW81 A0A2J7PX03 A0A2J7RL19 A0A1W7RAL1 A0A2J7PZJ9 A0A2J7PQA3 A0A2S2PMS6 A0A2J7RLI7 A0A2J7PMJ3 A0A2J7Q2Z3 A0A2J7Q5D5 A0A2J7RP47 A0A2V9X8L4 A0A2J7QGT4 A0A2J7RGX2 A0A2J7Q7C6 A0A2J7Q6A8 A0A2J7Q0C6 A0A2J7QRF9 A0A2J7QH51 A0A2J7QKB0 A0A2J7R4R4 A0A2J7QUN7 A0A3Q1M6J7 A0A2J7Q3T8 A0A2J7QGJ2 A0A2J7PHM9 A0A3Q1LX64 A0A2J7PTS1 A0A2J7R2D1 A0A2J7QSW0 A0A2J7Q647 A0A2J7Q0P9 A0A2J7QYI2 A0A2J7RJ94 A0A2J7RQT0 A0A2J7Q5X4
A0A2J7RLC6 A0A2J7PHX9 A0A2J7RNM5 A0A2J7QMH0 A0A2J7PJ38 A0A2J7R3B7 A0A2J7R639 A0A2J7R7D2 A0A2J7QLX7 A0A2J7R696 A0A2J7QQQ3 A0A2J7R4W1 A0A2J7RLG7 A0A2J7RRE4 A0A2J7PJ03 A0A2J7PF68 A0A2J7QNQ3 A0A2J7PZN7 A0A2J7RBL5 A0A2J7PBF1 A0A2J7RBL1 A0A2J7QSZ5 A0A2J7Q649 A0A2J7RPX5 A0A2J7PC89 A0A2J7RAC6 A0A2J7PQH3 A0A2J7R8W8 A0A2J7QJ13 A0A2J7QCG2 A0A2J7QEJ0 A0A2J7RBD8 A0A2J7RBX3 A0A2J7Q2Y3 A0A2J7PY33 A0A2J7Q2T8 A0A2J7PVW0 A0A2J7PT27 A0A2J7R8P5 A0A2J7R901 A0A2J7QUD0 A0A2J7R8G1 A0A2J7QJJ9 A0A2J7QMC5 A0A2J7QI56 A0A2J7Q6P7 A0A2J7RIL8 A0A2J7PNH4 A0A2J7PWV5 A0A3Q1LQN1 A0A2J7REE7 A0A2J7Q3K9 A0A2J7QCG0 A0A2J7QGX8 A0A2J7PTY0 A0A2J7QR01 A0A2J7PW81 A0A2J7PX03 A0A2J7RL19 A0A1W7RAL1 A0A2J7PZJ9 A0A2J7PQA3 A0A2S2PMS6 A0A2J7RLI7 A0A2J7PMJ3 A0A2J7Q2Z3 A0A2J7Q5D5 A0A2J7RP47 A0A2V9X8L4 A0A2J7QGT4 A0A2J7RGX2 A0A2J7Q7C6 A0A2J7Q6A8 A0A2J7Q0C6 A0A2J7QRF9 A0A2J7QH51 A0A2J7QKB0 A0A2J7R4R4 A0A2J7QUN7 A0A3Q1M6J7 A0A2J7Q3T8 A0A2J7QGJ2 A0A2J7PHM9 A0A3Q1LX64 A0A2J7PTS1 A0A2J7R2D1 A0A2J7QSW0 A0A2J7Q647 A0A2J7Q0P9 A0A2J7QYI2 A0A2J7RJ94 A0A2J7RQT0 A0A2J7Q5X4
PDB
5HHL
E-value=3.78979e-12,
Score=175
Ontologies
KEGG
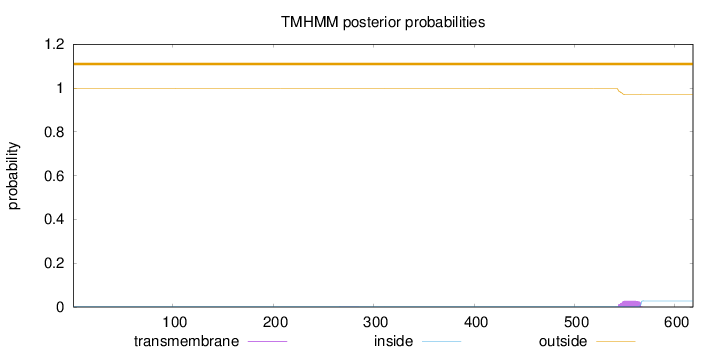
Topology
Length:
618
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.589349999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00320
outside
1 - 618
Population Genetic Test Statistics
Pi
305.98632
Theta
154.448114
Tajima's D
2.163007
CLR
56.359699
CSRT
0.906054697265137
Interpretation
Uncertain
