Pre Gene Modal
BGIBMGA009929
Annotation
PREDICTED:_atrophin-1-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.354
Sequence
CDS
ATGACGGCTTGGGGTACAACGGGCCAAAGGAAACGCATGAAAATACCCAGTGTCAAGTTGACGAAAAAATTTACTACCAAAGACAACAGACCTTTAATATACGAAACTGAGGATTTTTTCAAAAAACTCGAGAATCAACTTAAAGCTTTGAACTTAGGCAAGCCTCCGAAGACCAAGAAAAATGACATTCCATCGTCACAGCGTTCGAAGAATGACAGAGAAGTAAACGCTGGCAGTGAAACTCTTCGCGACGATGTAGATGATGAAGGTGGTGGAGAACAAAAAAGACGCGGCAAACGTCGTCGCAAAGGACGTCCAGGAGGCAGCGACCGCCTGACTTCGGCGGGATTGGCAGCTCAAACACAGCAAGACCCTGAAACACAAATCGCCGGAATAGGAACTGACTCACAGAACCCTAGTTCCCGAGGCTCCGTTGCTTCGACAGCCGATGATGATATCATTGTACCGCCGGTTGTCAAGACCAGTATAGCGAAGAAGACCGACTCATTTCTCGATGACGATATATTGAAATACCTGCACAGAGAAGTCGATGAAGAAGCAATCGAAATTGAATTTGATGTAAAGAGACGTTACGTCCTTGAAGAAGCTCTGCGTACGCGTCCCGATCGGCCTCAAGGTCAAGAGATGCAGGCTTTACTTCGGGAACTGAAGGTACCAGCTGTAAGTCTCGGGGACTGGCTACACATACCAAGAGTGTTCTCGAGACAGAACGCACAGTTTTCATTGCCGATAGATTCGAATGAACTTGAGGTTTTAACTCCAATGAACTACGCAGCGAGATTCGTATCTTTGAAAAAGTCAAAGCAATTATTGTACCTGACCGTGTTGAGGAAATTTCGTCCGAACGGCTATAGGATGCCTATAAAGGATATAGAAGATGGTCTTATCTTAATGATGGGTGGTATTATGACACTAAGTCAAGTCAACCACTTCAAAGAAATCATGAGCTGGACGTATTATGAGGAAGAAGAGAGTATACCGGACGAAATTAAGTTGACCTTACTGAACACCGGATACAGGAAGAACTCTGTTTCTGAGGGAGGAGATTCAGAAGAGCCTGAAGGACACACAGTTAAATTTAGAACTTGGTGTGGTCTGTGCGCGATTTGTGAACGCATGTACGGACGATTTCCGCTTCGAGAGAAAGATCCCCCAGACGGGATTGAATTAAGTGATTTTAGCATGATAGAAGTTAAACTAGCTACACTTAAAGTAAACAGTGGGTTGAGAGAAATTTTAAATACAATAAAAGAACGATAA
Protein
MTAWGTTGQRKRMKIPSVKLTKKFTTKDNRPLIYETEDFFKKLENQLKALNLGKPPKTKKNDIPSSQRSKNDREVNAGSETLRDDVDDEGGGEQKRRGKRRRKGRPGGSDRLTSAGLAAQTQQDPETQIAGIGTDSQNPSSRGSVASTADDDIIVPPVVKTSIAKKTDSFLDDDILKYLHREVDEEAIEIEFDVKRRYVLEEALRTRPDRPQGQEMQALLRELKVPAVSLGDWLHIPRVFSRQNAQFSLPIDSNELEVLTPMNYAARFVSLKKSKQLLYLTVLRKFRPNGYRMPIKDIEDGLILMMGGIMTLSQVNHFKEIMSWTYYEEEESIPDEIKLTLLNTGYRKNSVSEGGDSEEPEGHTVKFRTWCGLCAICERMYGRFPLREKDPPDGIELSDFSMIEVKLATLKVNSGLREILNTIKER
Summary
Uniprot
H9JK78
A0A2H1VPW1
A0A2A4JB63
A0A3S2NRA0
A0A194PQW4
A0A0N1PHK7
+ More
D6WXC6 A0A1B6G704 N6SYI2 E0V9K6 A0A2J7PU60 A0A2J7PU52 A0A2P8Z4Q3 A0A0C9RZU0 A0A067RH25 A0A0C9R853 A0A2A3EMF3 F4X6U9 A0A088A4W9 A0A087ZR69 A0A195CI52 A0A151XC43 A0A2S2NQJ6 A0A3Q0JKQ4 A0A195E7P4 E1ZYY2 K7IV40 A0A158NGC8 J9JYT2 A0A0M8ZXJ4 A0A154NZT6 A0A1B0CHW1 A0A182GXZ8 D1MLM5 A0A182HD51 A0A310SCK3 A0A1S4FQA8 A0A2P8Z0N2 A0A182JPR6 A0A084W4F2 Q16T82 A0A182W265 B0X4P1 A0A182M4X5 A0A182N9Q6 A0A182PK99 A0A0L7QJS2 A0A336LZR8 A0A182RMR2 A0A182QF22 A0A182IRK7 A0A1W4VN97 Q7PV52 W5JR65 A0A182FU91 A0A182UYA0 A0A1S4H925 A0A182XFR0 B3NDK3 A0A182HHS8 A0A182KLM5 A0A182TW65 B3MAC5 A0A0M5IZ21 A0A0M4EIU9 A0A182SVD6 A0A0L0CAP5 B4JZZ7 B4MX69 A0A182YBQ9 B4L0I4 Q2M1D8 B4GU95 A0A1I8M4I6 A0A3B0K6H6 Q9VUR8 A0A1I8Q2R4 B4QL46 B4HIH4 B4LII6 B4PCJ4 B4PCH6
D6WXC6 A0A1B6G704 N6SYI2 E0V9K6 A0A2J7PU60 A0A2J7PU52 A0A2P8Z4Q3 A0A0C9RZU0 A0A067RH25 A0A0C9R853 A0A2A3EMF3 F4X6U9 A0A088A4W9 A0A087ZR69 A0A195CI52 A0A151XC43 A0A2S2NQJ6 A0A3Q0JKQ4 A0A195E7P4 E1ZYY2 K7IV40 A0A158NGC8 J9JYT2 A0A0M8ZXJ4 A0A154NZT6 A0A1B0CHW1 A0A182GXZ8 D1MLM5 A0A182HD51 A0A310SCK3 A0A1S4FQA8 A0A2P8Z0N2 A0A182JPR6 A0A084W4F2 Q16T82 A0A182W265 B0X4P1 A0A182M4X5 A0A182N9Q6 A0A182PK99 A0A0L7QJS2 A0A336LZR8 A0A182RMR2 A0A182QF22 A0A182IRK7 A0A1W4VN97 Q7PV52 W5JR65 A0A182FU91 A0A182UYA0 A0A1S4H925 A0A182XFR0 B3NDK3 A0A182HHS8 A0A182KLM5 A0A182TW65 B3MAC5 A0A0M5IZ21 A0A0M4EIU9 A0A182SVD6 A0A0L0CAP5 B4JZZ7 B4MX69 A0A182YBQ9 B4L0I4 Q2M1D8 B4GU95 A0A1I8M4I6 A0A3B0K6H6 Q9VUR8 A0A1I8Q2R4 B4QL46 B4HIH4 B4LII6 B4PCJ4 B4PCH6
Pubmed
19121390
26354079
18362917
19820115
23537049
20566863
+ More
29403074 24845553 21719571 20798317 20075255 21347285 26483478 20920202 24438588 17510324 12364791 20920257 23761445 17994087 20966253 26108605 25244985 15632085 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17550304
29403074 24845553 21719571 20798317 20075255 21347285 26483478 20920202 24438588 17510324 12364791 20920257 23761445 17994087 20966253 26108605 25244985 15632085 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17550304
EMBL
BABH01039550
BABH01039551
BABH01039552
ODYU01003732
SOQ42883.1
NWSH01002166
+ More
PCG69006.1 RSAL01000127 RVE46540.1 KQ459595 KPI95702.1 KQ459784 KPJ19937.1 KQ971361 EFA08824.1 GECZ01011559 JAS58210.1 APGK01052113 KB741209 ENN72844.1 DS234994 EEB10062.1 NEVH01021208 PNF19870.1 PNF19867.1 PYGN01000196 PSN51473.1 GBYB01013656 JAG83423.1 KK852473 KDR23116.1 GBYB01009062 JAG78829.1 KZ288219 PBC32221.1 GL888818 EGI57881.1 KQ977754 KYN00107.1 KQ982314 KYQ57925.1 GGMR01006836 MBY19455.1 KQ979568 KYN20867.1 GL435242 EFN73672.1 AAZX01011129 ADTU01014817 ABLF02026693 KQ435833 KOX71623.1 KQ434782 KZC04508.1 AJWK01012770 AJWK01012771 AJWK01012772 JXUM01096545 KQ564272 KXJ72495.1 GU196316 ACZ26276.1 JXUM01033969 JXUM01033970 KQ561007 KXJ80060.1 KQ763450 OAD55044.1 PYGN01000253 PSN50040.1 ATLV01020330 KE525298 KFB45096.1 CH477658 EAT37672.1 DS232349 EDS40458.1 AXCM01001492 KQ415047 KOC58853.1 UFQT01000062 SSX19198.1 AXCN02001803 AAAB01008986 EAA00446.5 ADMH02000726 ETN65239.1 CH954178 EDV52136.2 APCN01002438 CH902618 EDV40176.2 CP012525 ALC44736.1 ALC44740.1 JRES01000760 KNC28524.1 CH916383 EDV98679.1 CH963876 EDW76902.1 CH933809 EDW19153.1 CH379069 EAL30636.2 CH479191 EDW26178.1 OUUW01000027 SPP89735.1 AE014296 AAF49607.2 CM000363 CM002912 EDX10571.1 KMY99791.1 CH480815 EDW41604.1 CH940647 EDW70773.2 CM000159 EDW94904.1 EDW94886.2
PCG69006.1 RSAL01000127 RVE46540.1 KQ459595 KPI95702.1 KQ459784 KPJ19937.1 KQ971361 EFA08824.1 GECZ01011559 JAS58210.1 APGK01052113 KB741209 ENN72844.1 DS234994 EEB10062.1 NEVH01021208 PNF19870.1 PNF19867.1 PYGN01000196 PSN51473.1 GBYB01013656 JAG83423.1 KK852473 KDR23116.1 GBYB01009062 JAG78829.1 KZ288219 PBC32221.1 GL888818 EGI57881.1 KQ977754 KYN00107.1 KQ982314 KYQ57925.1 GGMR01006836 MBY19455.1 KQ979568 KYN20867.1 GL435242 EFN73672.1 AAZX01011129 ADTU01014817 ABLF02026693 KQ435833 KOX71623.1 KQ434782 KZC04508.1 AJWK01012770 AJWK01012771 AJWK01012772 JXUM01096545 KQ564272 KXJ72495.1 GU196316 ACZ26276.1 JXUM01033969 JXUM01033970 KQ561007 KXJ80060.1 KQ763450 OAD55044.1 PYGN01000253 PSN50040.1 ATLV01020330 KE525298 KFB45096.1 CH477658 EAT37672.1 DS232349 EDS40458.1 AXCM01001492 KQ415047 KOC58853.1 UFQT01000062 SSX19198.1 AXCN02001803 AAAB01008986 EAA00446.5 ADMH02000726 ETN65239.1 CH954178 EDV52136.2 APCN01002438 CH902618 EDV40176.2 CP012525 ALC44736.1 ALC44740.1 JRES01000760 KNC28524.1 CH916383 EDV98679.1 CH963876 EDW76902.1 CH933809 EDW19153.1 CH379069 EAL30636.2 CH479191 EDW26178.1 OUUW01000027 SPP89735.1 AE014296 AAF49607.2 CM000363 CM002912 EDX10571.1 KMY99791.1 CH480815 EDW41604.1 CH940647 EDW70773.2 CM000159 EDW94904.1 EDW94886.2
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007266
+ More
UP000019118 UP000009046 UP000235965 UP000245037 UP000027135 UP000242457 UP000007755 UP000005203 UP000078542 UP000075809 UP000079169 UP000078492 UP000000311 UP000002358 UP000005205 UP000007819 UP000053105 UP000076502 UP000092461 UP000069940 UP000249989 UP000075881 UP000030765 UP000008820 UP000075920 UP000002320 UP000075883 UP000075884 UP000075885 UP000053825 UP000075900 UP000075886 UP000075880 UP000192221 UP000007062 UP000000673 UP000069272 UP000075903 UP000076407 UP000008711 UP000075840 UP000075882 UP000075902 UP000007801 UP000092553 UP000075901 UP000037069 UP000001070 UP000007798 UP000076408 UP000009192 UP000001819 UP000008744 UP000095301 UP000268350 UP000000803 UP000095300 UP000000304 UP000001292 UP000008792 UP000002282
UP000019118 UP000009046 UP000235965 UP000245037 UP000027135 UP000242457 UP000007755 UP000005203 UP000078542 UP000075809 UP000079169 UP000078492 UP000000311 UP000002358 UP000005205 UP000007819 UP000053105 UP000076502 UP000092461 UP000069940 UP000249989 UP000075881 UP000030765 UP000008820 UP000075920 UP000002320 UP000075883 UP000075884 UP000075885 UP000053825 UP000075900 UP000075886 UP000075880 UP000192221 UP000007062 UP000000673 UP000069272 UP000075903 UP000076407 UP000008711 UP000075840 UP000075882 UP000075902 UP000007801 UP000092553 UP000075901 UP000037069 UP000001070 UP000007798 UP000076408 UP000009192 UP000001819 UP000008744 UP000095301 UP000268350 UP000000803 UP000095300 UP000000304 UP000001292 UP000008792 UP000002282
Pfam
Interpro
IPR002951
Atrophin-like
+ More
IPR017884 SANT_dom
IPR009057 Homeobox-like_sf
IPR001005 SANT/Myb
IPR000949 ELM2_dom
IPR040007 Tho2
IPR021418 THO_THOC2_C
IPR032302 THOC2_N
IPR021726 THO_THOC2_N
IPR012476 GLE1
IPR038506 GLE1-like_sf
IPR001200 Phosducin
IPR036249 Thioredoxin-like_sf
IPR023196 Phosducin_N_dom_sf
IPR024253 Phosducin_thioredoxin-like_dom
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR023142 MAST_pre-PK_dom_sf
IPR015022 MA_Ser/Thr_Kinase_dom
IPR000719 Prot_kinase_dom
IPR017884 SANT_dom
IPR009057 Homeobox-like_sf
IPR001005 SANT/Myb
IPR000949 ELM2_dom
IPR040007 Tho2
IPR021418 THO_THOC2_C
IPR032302 THOC2_N
IPR021726 THO_THOC2_N
IPR012476 GLE1
IPR038506 GLE1-like_sf
IPR001200 Phosducin
IPR036249 Thioredoxin-like_sf
IPR023196 Phosducin_N_dom_sf
IPR024253 Phosducin_thioredoxin-like_dom
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR023142 MAST_pre-PK_dom_sf
IPR015022 MA_Ser/Thr_Kinase_dom
IPR000719 Prot_kinase_dom
Gene 3D
ProteinModelPortal
H9JK78
A0A2H1VPW1
A0A2A4JB63
A0A3S2NRA0
A0A194PQW4
A0A0N1PHK7
+ More
D6WXC6 A0A1B6G704 N6SYI2 E0V9K6 A0A2J7PU60 A0A2J7PU52 A0A2P8Z4Q3 A0A0C9RZU0 A0A067RH25 A0A0C9R853 A0A2A3EMF3 F4X6U9 A0A088A4W9 A0A087ZR69 A0A195CI52 A0A151XC43 A0A2S2NQJ6 A0A3Q0JKQ4 A0A195E7P4 E1ZYY2 K7IV40 A0A158NGC8 J9JYT2 A0A0M8ZXJ4 A0A154NZT6 A0A1B0CHW1 A0A182GXZ8 D1MLM5 A0A182HD51 A0A310SCK3 A0A1S4FQA8 A0A2P8Z0N2 A0A182JPR6 A0A084W4F2 Q16T82 A0A182W265 B0X4P1 A0A182M4X5 A0A182N9Q6 A0A182PK99 A0A0L7QJS2 A0A336LZR8 A0A182RMR2 A0A182QF22 A0A182IRK7 A0A1W4VN97 Q7PV52 W5JR65 A0A182FU91 A0A182UYA0 A0A1S4H925 A0A182XFR0 B3NDK3 A0A182HHS8 A0A182KLM5 A0A182TW65 B3MAC5 A0A0M5IZ21 A0A0M4EIU9 A0A182SVD6 A0A0L0CAP5 B4JZZ7 B4MX69 A0A182YBQ9 B4L0I4 Q2M1D8 B4GU95 A0A1I8M4I6 A0A3B0K6H6 Q9VUR8 A0A1I8Q2R4 B4QL46 B4HIH4 B4LII6 B4PCJ4 B4PCH6
D6WXC6 A0A1B6G704 N6SYI2 E0V9K6 A0A2J7PU60 A0A2J7PU52 A0A2P8Z4Q3 A0A0C9RZU0 A0A067RH25 A0A0C9R853 A0A2A3EMF3 F4X6U9 A0A088A4W9 A0A087ZR69 A0A195CI52 A0A151XC43 A0A2S2NQJ6 A0A3Q0JKQ4 A0A195E7P4 E1ZYY2 K7IV40 A0A158NGC8 J9JYT2 A0A0M8ZXJ4 A0A154NZT6 A0A1B0CHW1 A0A182GXZ8 D1MLM5 A0A182HD51 A0A310SCK3 A0A1S4FQA8 A0A2P8Z0N2 A0A182JPR6 A0A084W4F2 Q16T82 A0A182W265 B0X4P1 A0A182M4X5 A0A182N9Q6 A0A182PK99 A0A0L7QJS2 A0A336LZR8 A0A182RMR2 A0A182QF22 A0A182IRK7 A0A1W4VN97 Q7PV52 W5JR65 A0A182FU91 A0A182UYA0 A0A1S4H925 A0A182XFR0 B3NDK3 A0A182HHS8 A0A182KLM5 A0A182TW65 B3MAC5 A0A0M5IZ21 A0A0M4EIU9 A0A182SVD6 A0A0L0CAP5 B4JZZ7 B4MX69 A0A182YBQ9 B4L0I4 Q2M1D8 B4GU95 A0A1I8M4I6 A0A3B0K6H6 Q9VUR8 A0A1I8Q2R4 B4QL46 B4HIH4 B4LII6 B4PCJ4 B4PCH6
Ontologies
GO
PANTHER
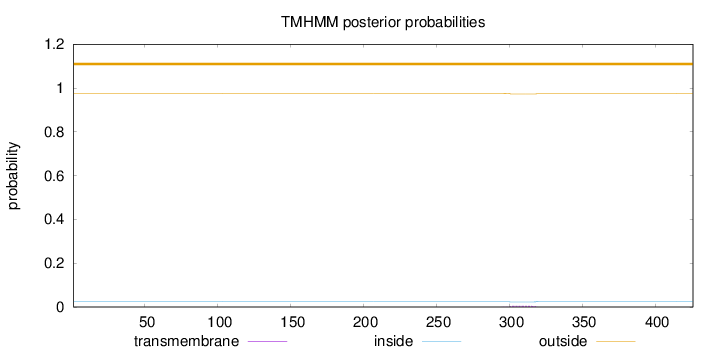
Topology
Subcellular location
Nucleus
Length:
426
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0887
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02404
outside
1 - 426
Population Genetic Test Statistics
Pi
275.058106
Theta
201.907452
Tajima's D
0.985161
CLR
0
CSRT
0.660516974151292
Interpretation
Uncertain
