Gene
KWMTBOMO04707
Pre Gene Modal
BGIBMGA009937
Annotation
PREDICTED:_ankyrin_repeat_and_zinc_finger_domain-containing_protein_1-like_isoform_X4_[Bombyx_mori]
Full name
Ankyrin repeat and zinc finger domain-containing protein 1
Location in the cell
Nuclear Reliability : 4.406
Sequence
CDS
ATGGCGCTGAAGCCCGAGGCAGTAAAAACTAAAATCGTGAAAGTCTATGAGATCGACGAATTTCAAAAACTCCTAAAAGGTGTTCATGTGGCAGCATGTATGATAACAGAATCACATAATGTCATTGATGAACAGAGTGTCTTGCGTCGCCTTAATGCTTTAACCTTAAATGGTGCTTCTGATGGTAATTGTTGTTCGTGTTGCGGAGTGGGTCCATTCGAGACCCGTGCGCAGCAAACGGCTCATTATAAGCATCACTGGCACACTTTTAACTTGAAGAGGAAACTATTTGGAAAATCTCCATTATCCATTGGCCAGTATAATTCTCGGAAAGATGAATCAAGCAGTGAGTCTGAGGAAGAAGTGGAGGACAGTAAGAGACCTCCGAGTGACCTATTTGCTGCAGCAACAAGACATTGCAAGGCGTTCTTCACTAATCATTATGGGCAGGTCTTCTGTATTTATAGATGTATCCTACATCATAAGAAGGAAGAATTATCAACGGATGGTGATGGCAGTGCCTGGGTAGAACGTGTGGAGCAGCTATTGATCCCAGGCTATCAAAGATGGGCAGTCCTGATGGTCTCGGGTGGCAGATTTGCTGGGGCAGTTTTCAGTGGTGGCACTATAGTTCTACACAAGACATTTCATTCATATGTCACAAGGAGAGGGCAAGGTCAAGCCCAGGCTACAAGAGATGCTCATCATGGACACGCGAGGTCTGCCGGTGCTAGCCTTAGAAGATATAATCAGGCGCAGTTTCTAGAGCATATCCAAGATATTATAAGTCAATGGACAGAAGACTTCAAAGGTTGTTCTCTAATATTATATCGGGCTGTAGGATCTTTAAATCAAGCTGCTATATTTGGAAAGAACTCCCCTATAAAGAAAGATGATCCAAGACTTAGAGTGCTACCTTTCCCTACTAGAAAACCTACATTTAAAGAAGTTCAGCGGGTTCATTCTACACTTGCTAGTCTAGAGGTTTACGATTCGTTGGAGTTGTTCCAAAAAGCACTGATAACATCGACAGCTAAACCAGCAAATAAAGCTACACTGGATCTTGGTAATGAAAATATTAAGAAAACAGCAAAGAAAAGTCCTAGTAAAACTATTGATAGAGCAAAATCAAGAGAACGCACTCCTAGAGAGCTACCAACCCAGTTGATGTCGAGCGAAGACGAAGGTCCCTGCTTCATAGACTCTGAGACACCTGTAGGTTGGATCAAAACACTATCGGAACAAAAAACCAGTGGGATTATTATTAATTCTAAAAAAGTTGTCCTAAACGATTGTGCTATTCAAGATAGGTCAACACCTAGTCCCTGCGAGAGCGAGAAGGAAGATTTAAAGCAGGAAGTAAAAGAGAAATATGAAACTGATAGGAGGAAAAAGATAAAAGATCCCGCTGTTCTTAACAAGCCCAGTCAAAAGATTCCTAATCCTATTAAGAAGATGTGGAAAATGATAATAGGAAATGAATTGAAATCACTGGAGACGATATTCGAAGGCTGGGAGGAAGAGAATCTGGCGGAGGCTTGTAACACTCGCGATCCTAGCGATGGCAACACTGCGCTACACAAAGCCGCCATTGCTGCGAAACCAGATATGGTCACTGAGATCCTGACTGCCGGTGGAGATCCGTGCATCAGGAACAATCAGCTGCAGACCCCGTACGCGGCCTCGCCACACCAGGAGACACGCCTCGCCTTCAGAATGTTCCAGGCTCAGTTTCCAGACAAATACAATTACGCTAAGTCGCACATTCCCGGCCCGGTGACCCCCGAGACTCTGGAACAAGAGAAAGAGAGGAAGCTACAACAGAAACGAGCTAAGAGGCAACGAGACAAGGAGAAGCAATTGGAGAAGATTAAAACCAACAAGTTCCTTCAGATGACCGATACCGAAAAGGCGAAGTCATCGGAGAAGCGTTGTTTTCTTTGTGGCTCAAATCTCCCGAAGAAACCGTTCGAGTACGAAGTGTACAGATTCTGCGCAGTGCAATGTCTCCAGAACCACAGAAACATTCGACCTCTAAACATGAGCGCGTGA
Protein
MALKPEAVKTKIVKVYEIDEFQKLLKGVHVAACMITESHNVIDEQSVLRRLNALTLNGASDGNCCSCCGVGPFETRAQQTAHYKHHWHTFNLKRKLFGKSPLSIGQYNSRKDESSSESEEEVEDSKRPPSDLFAAATRHCKAFFTNHYGQVFCIYRCILHHKKEELSTDGDGSAWVERVEQLLIPGYQRWAVLMVSGGRFAGAVFSGGTIVLHKTFHSYVTRRGQGQAQATRDAHHGHARSAGASLRRYNQAQFLEHIQDIISQWTEDFKGCSLILYRAVGSLNQAAIFGKNSPIKKDDPRLRVLPFPTRKPTFKEVQRVHSTLASLEVYDSLELFQKALITSTAKPANKATLDLGNENIKKTAKKSPSKTIDRAKSRERTPRELPTQLMSSEDEGPCFIDSETPVGWIKTLSEQKTSGIIINSKKVVLNDCAIQDRSTPSPCESEKEDLKQEVKEKYETDRRKKIKDPAVLNKPSQKIPNPIKKMWKMIIGNELKSLETIFEGWEEENLAEACNTRDPSDGNTALHKAAIAAKPDMVTEILTAGGDPCIRNNQLQTPYAASPHQETRLAFRMFQAQFPDKYNYAKSHIPGPVTPETLEQEKERKLQQKRAKRQRDKEKQLEKIKTNKFLQMTDTEKAKSSEKRCFLCGSNLPKKPFEYEVYRFCAVQCLQNHRNIRPLNMSA
Summary
Description
Plays a role in the cellular response to hydrogen peroxide and in the maintenance of mitochondrial integrity under conditions of cellular stress (By similarity). Involved in the endoplasmic reticulum (ER)-associated degradation (ERAD) pathway (By similarity).
Subunit
Interacts (via VIM motif) with VCP.
Similarity
Belongs to the ANKZF1/VMS1 family.
Keywords
ANK repeat
Coiled coil
Complete proteome
Cytoplasm
Metal-binding
Phosphoprotein
Reference proteome
Repeat
Zinc
Zinc-finger
Feature
chain Ankyrin repeat and zinc finger domain-containing protein 1
Uniprot
H9JK86
A0A3S2P2V4
A0A194PRW1
A0A2A4JGB5
A0A0N1PJC0
A0A212EX95
+ More
A0A2H1VA90 A0A195EGX9 A0A224XL02 A0A069DZF8 A0A0A9VX70 A0A2J7RI97 A0A1Y1NCG2 A0A088A054 A0A2A3E523 A0A154P3C1 A0A2A3E4A7 A0A0L7R757 N6TMS3 A0A195F7K7 A0A1S4EN03 A0A151I4F0 A0A158NI24 A0A195CKK8 J9JXP6 A0A026W8N7 A0A151WJS7 E2C8J3 E2APC5 A0A0C9QF49 A0A0A9W064 A0A0N0BDV2 A0A1S3IMI7 A0A1S3ILJ1 A0A0J7L5I0 A0A2S2R626 H2THP4 A0A2U9C979 E7F4D7 A0A3S2PCV0 A0A1Y1NAM8 I3JR47 A0A2S2P475 A0A3P9GZQ1 A0A3P9BKM8 A0A3Q2UW42 A0A3P8Q0C5 A0A2R2MR32 A0A3B4EU68 A0A1D2M511 A0A0N7ZA02 A0A3Q2V4B2 F7A9G2 A0A226EWR2 G5AKK1 A0A0P4VQR9 A0A0N8ETG3 A0A2I3RLX9 A0A2R9AQV1 A0A2I2YQS1 F6WVX7 A0A1W2W4B3 G3HWF2 A0A2J8TUH4 Q66H85
A0A2H1VA90 A0A195EGX9 A0A224XL02 A0A069DZF8 A0A0A9VX70 A0A2J7RI97 A0A1Y1NCG2 A0A088A054 A0A2A3E523 A0A154P3C1 A0A2A3E4A7 A0A0L7R757 N6TMS3 A0A195F7K7 A0A1S4EN03 A0A151I4F0 A0A158NI24 A0A195CKK8 J9JXP6 A0A026W8N7 A0A151WJS7 E2C8J3 E2APC5 A0A0C9QF49 A0A0A9W064 A0A0N0BDV2 A0A1S3IMI7 A0A1S3ILJ1 A0A0J7L5I0 A0A2S2R626 H2THP4 A0A2U9C979 E7F4D7 A0A3S2PCV0 A0A1Y1NAM8 I3JR47 A0A2S2P475 A0A3P9GZQ1 A0A3P9BKM8 A0A3Q2UW42 A0A3P8Q0C5 A0A2R2MR32 A0A3B4EU68 A0A1D2M511 A0A0N7ZA02 A0A3Q2V4B2 F7A9G2 A0A226EWR2 G5AKK1 A0A0P4VQR9 A0A0N8ETG3 A0A2I3RLX9 A0A2R9AQV1 A0A2I2YQS1 F6WVX7 A0A1W2W4B3 G3HWF2 A0A2J8TUH4 Q66H85
Pubmed
EMBL
BABH01039520
RSAL01000041
RVE50938.1
KQ459595
KPI95693.1
NWSH01001561
+ More
PCG70859.1 KQ459784 KPJ19926.1 AGBW02011805 OWR46109.1 ODYU01001469 SOQ37706.1 KQ978957 KYN27142.1 GFTR01007717 JAW08709.1 GBGD01000715 JAC88174.1 GBHO01042797 GBRD01013116 GDHC01019157 JAG00807.1 JAG52710.1 JAP99471.1 NEVH01003502 PNF40559.1 GEZM01009457 GEZM01009456 JAV94580.1 KZ288379 PBC26594.1 KQ434809 KZC06435.1 PBC26593.1 KQ414643 KOC66669.1 APGK01058603 KB741291 KB632343 ENN70520.1 ERL93018.1 KQ981763 KYN36049.1 KQ976456 KYM84879.1 ADTU01016189 KQ977622 KYN01265.1 ABLF02003796 KK107341 QOIP01000012 EZA52420.1 RLU16320.1 KQ983039 KYQ48057.1 GL453666 EFN75723.1 GL441504 EFN64724.1 GBYB01002004 JAG71771.1 GBHO01042798 JAG00806.1 KQ435848 KOX70984.1 LBMM01000688 KMQ97754.1 GGMS01015609 MBY84812.1 CP026256 AWP13018.1 CR752651 CM012457 RVE58082.1 GEZM01009458 GEZM01009455 JAV94578.1 AERX01017599 GGMR01011644 MBY24263.1 LJIJ01004272 ODM88001.1 GDRN01107043 JAI57480.1 AAMC01028606 AAMC01028607 LNIX01000001 OXA61504.1 JH165659 EHA97561.1 GDRN01107050 JAI57479.1 GEBF01004459 JAN99173.1 AACZ04056567 AJFE02063776 EAAA01000312 JH000831 EGW11868.1 NDHI03003483 PNJ36662.1 BC081973
PCG70859.1 KQ459784 KPJ19926.1 AGBW02011805 OWR46109.1 ODYU01001469 SOQ37706.1 KQ978957 KYN27142.1 GFTR01007717 JAW08709.1 GBGD01000715 JAC88174.1 GBHO01042797 GBRD01013116 GDHC01019157 JAG00807.1 JAG52710.1 JAP99471.1 NEVH01003502 PNF40559.1 GEZM01009457 GEZM01009456 JAV94580.1 KZ288379 PBC26594.1 KQ434809 KZC06435.1 PBC26593.1 KQ414643 KOC66669.1 APGK01058603 KB741291 KB632343 ENN70520.1 ERL93018.1 KQ981763 KYN36049.1 KQ976456 KYM84879.1 ADTU01016189 KQ977622 KYN01265.1 ABLF02003796 KK107341 QOIP01000012 EZA52420.1 RLU16320.1 KQ983039 KYQ48057.1 GL453666 EFN75723.1 GL441504 EFN64724.1 GBYB01002004 JAG71771.1 GBHO01042798 JAG00806.1 KQ435848 KOX70984.1 LBMM01000688 KMQ97754.1 GGMS01015609 MBY84812.1 CP026256 AWP13018.1 CR752651 CM012457 RVE58082.1 GEZM01009458 GEZM01009455 JAV94578.1 AERX01017599 GGMR01011644 MBY24263.1 LJIJ01004272 ODM88001.1 GDRN01107043 JAI57480.1 AAMC01028606 AAMC01028607 LNIX01000001 OXA61504.1 JH165659 EHA97561.1 GDRN01107050 JAI57479.1 GEBF01004459 JAN99173.1 AACZ04056567 AJFE02063776 EAAA01000312 JH000831 EGW11868.1 NDHI03003483 PNJ36662.1 BC081973
Proteomes
UP000005204
UP000283053
UP000053268
UP000218220
UP000053240
UP000007151
+ More
UP000078492 UP000235965 UP000005203 UP000242457 UP000076502 UP000053825 UP000019118 UP000030742 UP000078541 UP000079169 UP000078540 UP000005205 UP000078542 UP000007819 UP000053097 UP000279307 UP000075809 UP000008237 UP000000311 UP000053105 UP000085678 UP000036403 UP000005226 UP000246464 UP000000437 UP000005207 UP000265200 UP000265160 UP000264840 UP000265100 UP000261460 UP000094527 UP000008143 UP000198287 UP000006813 UP000002277 UP000240080 UP000001519 UP000008144 UP000001075 UP000002494
UP000078492 UP000235965 UP000005203 UP000242457 UP000076502 UP000053825 UP000019118 UP000030742 UP000078541 UP000079169 UP000078540 UP000005205 UP000078542 UP000007819 UP000053097 UP000279307 UP000075809 UP000008237 UP000000311 UP000053105 UP000085678 UP000036403 UP000005226 UP000246464 UP000000437 UP000005207 UP000265200 UP000265160 UP000264840 UP000265100 UP000261460 UP000094527 UP000008143 UP000198287 UP000006813 UP000002277 UP000240080 UP000001519 UP000008144 UP000001075 UP000002494
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JK86
A0A3S2P2V4
A0A194PRW1
A0A2A4JGB5
A0A0N1PJC0
A0A212EX95
+ More
A0A2H1VA90 A0A195EGX9 A0A224XL02 A0A069DZF8 A0A0A9VX70 A0A2J7RI97 A0A1Y1NCG2 A0A088A054 A0A2A3E523 A0A154P3C1 A0A2A3E4A7 A0A0L7R757 N6TMS3 A0A195F7K7 A0A1S4EN03 A0A151I4F0 A0A158NI24 A0A195CKK8 J9JXP6 A0A026W8N7 A0A151WJS7 E2C8J3 E2APC5 A0A0C9QF49 A0A0A9W064 A0A0N0BDV2 A0A1S3IMI7 A0A1S3ILJ1 A0A0J7L5I0 A0A2S2R626 H2THP4 A0A2U9C979 E7F4D7 A0A3S2PCV0 A0A1Y1NAM8 I3JR47 A0A2S2P475 A0A3P9GZQ1 A0A3P9BKM8 A0A3Q2UW42 A0A3P8Q0C5 A0A2R2MR32 A0A3B4EU68 A0A1D2M511 A0A0N7ZA02 A0A3Q2V4B2 F7A9G2 A0A226EWR2 G5AKK1 A0A0P4VQR9 A0A0N8ETG3 A0A2I3RLX9 A0A2R9AQV1 A0A2I2YQS1 F6WVX7 A0A1W2W4B3 G3HWF2 A0A2J8TUH4 Q66H85
A0A2H1VA90 A0A195EGX9 A0A224XL02 A0A069DZF8 A0A0A9VX70 A0A2J7RI97 A0A1Y1NCG2 A0A088A054 A0A2A3E523 A0A154P3C1 A0A2A3E4A7 A0A0L7R757 N6TMS3 A0A195F7K7 A0A1S4EN03 A0A151I4F0 A0A158NI24 A0A195CKK8 J9JXP6 A0A026W8N7 A0A151WJS7 E2C8J3 E2APC5 A0A0C9QF49 A0A0A9W064 A0A0N0BDV2 A0A1S3IMI7 A0A1S3ILJ1 A0A0J7L5I0 A0A2S2R626 H2THP4 A0A2U9C979 E7F4D7 A0A3S2PCV0 A0A1Y1NAM8 I3JR47 A0A2S2P475 A0A3P9GZQ1 A0A3P9BKM8 A0A3Q2UW42 A0A3P8Q0C5 A0A2R2MR32 A0A3B4EU68 A0A1D2M511 A0A0N7ZA02 A0A3Q2V4B2 F7A9G2 A0A226EWR2 G5AKK1 A0A0P4VQR9 A0A0N8ETG3 A0A2I3RLX9 A0A2R9AQV1 A0A2I2YQS1 F6WVX7 A0A1W2W4B3 G3HWF2 A0A2J8TUH4 Q66H85
PDB
5WHG
E-value=2.74504e-15,
Score=202
Ontologies
KEGG
GO
Topology
Subcellular location
Cytoplasm
Translocates to the mitochondria upon exposure to hydrogen peroxide. With evidence from 1 publications.
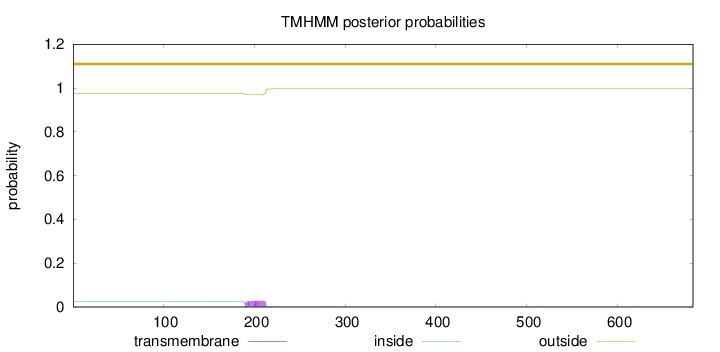
Length:
683
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.63517
Exp number, first 60 AAs:
0.0008
Total prob of N-in:
0.02552
outside
1 - 683
Population Genetic Test Statistics
Pi
246.67667
Theta
180.132644
Tajima's D
1.124163
CLR
0.13387
CSRT
0.692165391730413
Interpretation
Uncertain
