Gene
KWMTBOMO04704
Pre Gene Modal
BGIBMGA009939
Annotation
PREDICTED:_hemicentin-1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.679 Nuclear Reliability : 1.146 PlasmaMembrane Reliability : 1.064
Sequence
CDS
ATGTATTTGCCCGTAGGCATCTACCGTGGTAAATATGAGTGGGCAAACGAGAACAGTCCCGTTGGAGGAGACTGCTCACTTTGGGTACGCGCTGCGACACTTCAGCTTGATGATGGTCAATGGCAGTGTCAAGTTACTGCCAGTAATTATGACGTACAGGACGCCTTATCAAGTCCACCAGCTCCTTTAGCAGTTCGAGTGCCTCCACAATCTCCGAGGATCCTATTCAACGGCTCCCACGTGCTACCAGGCCAAAACATCACAGTTCCAGCGGGGTCCAGAGCGACAGTCGTCTGTGAAGCCAGATATGGCAATCCACCTGCATATATCGAGTGGTATTTAGAAAAAGAACGCCTGACAGCGTGGAGCCAAACCAATGCTTCGGAAGTGGAGCGACCTCGGGTTTGGGCGGCCAGATCCGTGTTGGAACTCGGAGCGACGCGTTCGGCTCACGGCCGGACTTTGGCTTGTAGGGCGCATCACCCTTCTTATCCGGCGCCGTACTACCGGGACTCCTACACGATGCTGGATGTCACTTATGCTCCTGTGGTTTCCATAAACGGCGCCGACGCGAGCACCCTGGCCAATCTGGAGGAAGGTTCCAGTGCTCTATCTCTGGAGTGCAGGGCCAACGGGAACCCTAGCCCTTACGTGTGGTGGACTAGGAACGGACAAGTCATACCCACAAATGGCCCAAAACTGATCCTGGCTCCTATAGTCAGGAATCATTCAGGTATCTATGGATGCCAAGCCCGAAATTCGGCCGGGACCTCCGACTCGGTGAAAATTGAAATTGATGTCAAATATCCACCGAGGGTCACTTGGGTCGGGCCGGACACAGTGGTGGAAGCCAATCTCTTCTCGCAAATTACTTTGGAGTGCAAGGCTGAGGGGAACCCGCCTCCTTCCTACCAGTGGTACCACAGCCCGGGGGGCGCCGCCCGCATCAACGACGCGCAGGAGGAGGGCGTCCCCGTGTCCTCCACGCCGCAGCTGCAGCTGCACAACGTGTCGTACGCGCAGCACGGCCGCTACACCTGCGTCGCCTCCAACCGCATCGGCCTTCAGGACAGGAGTCACCAGTCGGAGGCGATCCGCGTGAACGTGTTGGGTCCGCCGGTGGGGGAGGCGCGGGGGGCGGCGCACGCGTGGCGGGGGGCCGCCGTCCGCCTGCACGCCGCCCTGTGCGCCGACCCGCCGCCGCGCCGCGCCGTCTGGACCTGGGGCAGTCTGCGCCTCGACGTGCCCGGGGAACTAGGTCGTTTTAGTTCGCTGGAACCGTCTTCCGAGAACGGCTGCTACCGGTACACGCTGCTCGTGTCCAATGTGGCGCCGGACGACGCCCGCGCCTACGTGCTGCGCGTCGAGAACGAGCGCGGCTCCTCCACGCACGCCGTGGCGCTCACGGTGCACGATAATTTTTCGCTAACAGAGACGTCGCACGCGGCCCCGCTCGCGGCCGCCGCGCTGGTCTCCGCCGCGCTGCTGGTGCTCGTGCTGTGCCTCGTCGTCAGGTGCAGGAGGAGGAGAGAAAGCGTCGAGTACAAGACTGATGACTTAAATAGCGAGAAGACCGTGCTCCCAGCGGACGCGGTGTACACGCCGCGCGTCCCTGCGAGCCCCCCCGTCCCCCCGAGCGTGGCGGGTCCCCCCGGCCCCCCGGGCGCGGCGGTCCCCCGAGCGGCGACGGGGGGCCGGTACTCCCCGGGCGCGCTGCAGGTCCGGCGCGCGGCCGTGGTGCTGCAGCCGCCCACCACCGTGTGA
Protein
MYLPVGIYRGKYEWANENSPVGGDCSLWVRAATLQLDDGQWQCQVTASNYDVQDALSSPPAPLAVRVPPQSPRILFNGSHVLPGQNITVPAGSRATVVCEARYGNPPAYIEWYLEKERLTAWSQTNASEVERPRVWAARSVLELGATRSAHGRTLACRAHHPSYPAPYYRDSYTMLDVTYAPVVSINGADASTLANLEEGSSALSLECRANGNPSPYVWWTRNGQVIPTNGPKLILAPIVRNHSGIYGCQARNSAGTSDSVKIEIDVKYPPRVTWVGPDTVVEANLFSQITLECKAEGNPPPSYQWYHSPGGAARINDAQEEGVPVSSTPQLQLHNVSYAQHGRYTCVASNRIGLQDRSHQSEAIRVNVLGPPVGEARGAAHAWRGAAVRLHAALCADPPPRRAVWTWGSLRLDVPGELGRFSSLEPSSENGCYRYTLLVSNVAPDDARAYVLRVENERGSSTHAVALTVHDNFSLTETSHAAPLAAAALVSAALLVLVLCLVVRCRRRRESVEYKTDDLNSEKTVLPADAVYTPRVPASPPVPPSVAGPPGPPGAAVPRAATGGRYSPGALQVRRAAVVLQPPTTV
Summary
Uniprot
A0A2H1V8C3
A0A3S2NLT4
H9JK88
A0A212FGA5
A0A194PQV4
A0A0N1IAT8
+ More
A0A194PSG0 A0A067QK27 A0A182I8N7 A0A182F9A9 A0A182W155 A0A1J1IH42 A0A0L0BPR8 A0A182MD52 A0A182GR80 A0A1I8M182 A0A139WLQ8 A0A084VEE9 B3LZK5 A0A0P9AP37 Q295P3 A0A0N8P1B3 A0A1Y9IZV1 A0A1I8QAT0 A0A0R3NJ28 A0A1W4VTB0 A0A3B0KH40 A0A1D2N0V7 A0A1W4VTZ4 A0A126GUS1 A0A126GUR3 Q9VHQ8 A0A0R3NP57 A0A0R3NQS8 A0A1W4VUN3 A0A1W4VGC4 B5X504 E0W270 A0A182QVU9 Q7QFT8 A0A182XX40 B4KD35 A0A1W4VGL5 B4N9A1 A0A0B4K609 A0A0R1E1N3 A0A0B4K642 A0A034WGA0 A0A1B0B5M4 A0A0A1XJT7 Q9VHR2 W8C0L2 W8B1Y8 B4LZM1 A0A226ETC0 A0A026W2F3 E2AQP1 F5HJC7 E9GS53 A0A0P4YQI9 A0A1B0AC74 A0A154PGV5 A0A0C9RPS0 A0A0C9QBZ9 A0A2A3EJ29 A0A158NE28 A0A088A2P4 A0A151XED5 A0A0L7RBK8 A0A3L8E0B7 A0A0K2VEP7 A0A2P2I5N8 A0A151IF28 A0A195DB68 A0A0C9R6P0 A0A0P4XRC3 A0A0P5BGU0 A0A0P5MWX0 A0A0P5YZD4 A0A0P5BDB4 A0A0P5LB48 A0A1J1IJ21 A0A0P5UTM1 A0A0P5AM18 A0A195BP15 A0A0P6EVR9 A0A0N8AS41 A0A0P5YHC0 A0A164Z592 A0A0P5S452 A0A0P5ICW1 A0A0P5EIR9 A0A0N8C6Z2 A0A0P6EVV5 A0A0P5BF28 A0A0N8EEH0
A0A194PSG0 A0A067QK27 A0A182I8N7 A0A182F9A9 A0A182W155 A0A1J1IH42 A0A0L0BPR8 A0A182MD52 A0A182GR80 A0A1I8M182 A0A139WLQ8 A0A084VEE9 B3LZK5 A0A0P9AP37 Q295P3 A0A0N8P1B3 A0A1Y9IZV1 A0A1I8QAT0 A0A0R3NJ28 A0A1W4VTB0 A0A3B0KH40 A0A1D2N0V7 A0A1W4VTZ4 A0A126GUS1 A0A126GUR3 Q9VHQ8 A0A0R3NP57 A0A0R3NQS8 A0A1W4VUN3 A0A1W4VGC4 B5X504 E0W270 A0A182QVU9 Q7QFT8 A0A182XX40 B4KD35 A0A1W4VGL5 B4N9A1 A0A0B4K609 A0A0R1E1N3 A0A0B4K642 A0A034WGA0 A0A1B0B5M4 A0A0A1XJT7 Q9VHR2 W8C0L2 W8B1Y8 B4LZM1 A0A226ETC0 A0A026W2F3 E2AQP1 F5HJC7 E9GS53 A0A0P4YQI9 A0A1B0AC74 A0A154PGV5 A0A0C9RPS0 A0A0C9QBZ9 A0A2A3EJ29 A0A158NE28 A0A088A2P4 A0A151XED5 A0A0L7RBK8 A0A3L8E0B7 A0A0K2VEP7 A0A2P2I5N8 A0A151IF28 A0A195DB68 A0A0C9R6P0 A0A0P4XRC3 A0A0P5BGU0 A0A0P5MWX0 A0A0P5YZD4 A0A0P5BDB4 A0A0P5LB48 A0A1J1IJ21 A0A0P5UTM1 A0A0P5AM18 A0A195BP15 A0A0P6EVR9 A0A0N8AS41 A0A0P5YHC0 A0A164Z592 A0A0P5S452 A0A0P5ICW1 A0A0P5EIR9 A0A0N8C6Z2 A0A0P6EVV5 A0A0P5BF28 A0A0N8EEH0
Pubmed
19121390
22118469
26354079
24845553
26108605
26483478
+ More
25315136 18362917 19820115 24438588 17994087 15632085 27289101 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20566863 12364791 14747013 17210077 25244985 17550304 25348373 25830018 24495485 24508170 20798317 21292972 21347285 30249741
25315136 18362917 19820115 24438588 17994087 15632085 27289101 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20566863 12364791 14747013 17210077 25244985 17550304 25348373 25830018 24495485 24508170 20798317 21292972 21347285 30249741
EMBL
ODYU01001181
SOQ37046.1
RSAL01000041
RVE50937.1
BABH01039507
BABH01039508
+ More
BABH01039509 AGBW02008693 OWR52757.1 KQ459595 KPI95692.1 KQ459784 KPJ19927.1 KPI95694.1 KK853366 KDR08118.1 APCN01003096 CVRI01000051 CRK99539.1 JRES01001565 KNC22052.1 AXCM01000674 JXUM01002001 JXUM01002002 JXUM01002003 JXUM01002004 JXUM01002005 JXUM01002006 JXUM01002007 JXUM01002008 JXUM01002009 KQ560131 KXJ84278.1 KQ971319 KYB28872.1 ATLV01012275 KE524778 KFB36343.1 CH902617 EDV41947.2 KPU79445.1 CM000070 EAL28665.4 KPU79444.1 KRT00935.1 OUUW01000008 SPP84411.1 LJIJ01000310 ODM98937.1 AE014297 ALI30541.1 ALI30540.1 AAF54243.4 KRT00933.1 KRT00934.1 BT046123 ACI46511.1 DS235874 EEB19726.1 AXCN02000345 AAAB01008844 EAA06064.4 CH933806 EDW13805.2 CH964232 EDW80534.2 AFH06307.1 CM000160 KRK03180.1 AFH06308.1 GAKP01005258 JAC53694.1 JXJN01008779 GBXI01002658 JAD11634.1 AAF54239.4 GAMC01011391 JAB95164.1 GAMC01011390 JAB95165.1 CH940650 EDW67160.2 LNIX01000002 OXA60769.1 KK107474 EZA50178.1 GL441804 EFN64230.1 EGK96387.1 EGK96388.1 GL732561 EFX77651.1 GDIP01234565 JAI88836.1 KQ434899 KZC11037.1 GBYB01010475 JAG80242.1 GBYB01011998 JAG81765.1 KZ288252 PBC31031.1 ADTU01012618 ADTU01012619 ADTU01012620 ADTU01012621 ADTU01012622 KQ982254 KYQ58732.1 KQ414617 KOC68131.1 QOIP01000002 RLU25619.1 HACA01031306 CDW48667.1 IACF01003718 LAB69329.1 KQ977830 KYM99406.1 KQ981082 KYN09659.1 GBYB01011999 JAG81766.1 GDIP01239226 GDIQ01174802 GDIQ01059790 JAI84175.1 JAK76923.1 GDIP01199822 JAJ23580.1 GDIP01168925 GDIQ01152317 JAJ54477.1 JAK99408.1 GDIP01052398 JAM51317.1 GDIP01186133 JAJ37269.1 GDIQ01172453 JAK79272.1 CRK99540.1 GDIP01109685 JAL94029.1 GDIP01197374 JAJ26028.1 KQ976424 KYM88257.1 GDIQ01057241 JAN37496.1 GDIQ01248331 GDIQ01090834 JAK03394.1 GDIP01057941 JAM45774.1 LRGB01000781 KZS15954.1 GDIQ01097072 JAL54654.1 GDIQ01219294 JAK32431.1 GDIP01239225 GDIQ01268974 GDIQ01237277 GDIQ01140514 GDIQ01130364 JAI84176.1 JAJ82750.1 GDIQ01108723 JAL43003.1 GDIQ01057240 JAN37497.1 GDIP01186132 JAJ37270.1 GDIQ01034708 JAN60029.1
BABH01039509 AGBW02008693 OWR52757.1 KQ459595 KPI95692.1 KQ459784 KPJ19927.1 KPI95694.1 KK853366 KDR08118.1 APCN01003096 CVRI01000051 CRK99539.1 JRES01001565 KNC22052.1 AXCM01000674 JXUM01002001 JXUM01002002 JXUM01002003 JXUM01002004 JXUM01002005 JXUM01002006 JXUM01002007 JXUM01002008 JXUM01002009 KQ560131 KXJ84278.1 KQ971319 KYB28872.1 ATLV01012275 KE524778 KFB36343.1 CH902617 EDV41947.2 KPU79445.1 CM000070 EAL28665.4 KPU79444.1 KRT00935.1 OUUW01000008 SPP84411.1 LJIJ01000310 ODM98937.1 AE014297 ALI30541.1 ALI30540.1 AAF54243.4 KRT00933.1 KRT00934.1 BT046123 ACI46511.1 DS235874 EEB19726.1 AXCN02000345 AAAB01008844 EAA06064.4 CH933806 EDW13805.2 CH964232 EDW80534.2 AFH06307.1 CM000160 KRK03180.1 AFH06308.1 GAKP01005258 JAC53694.1 JXJN01008779 GBXI01002658 JAD11634.1 AAF54239.4 GAMC01011391 JAB95164.1 GAMC01011390 JAB95165.1 CH940650 EDW67160.2 LNIX01000002 OXA60769.1 KK107474 EZA50178.1 GL441804 EFN64230.1 EGK96387.1 EGK96388.1 GL732561 EFX77651.1 GDIP01234565 JAI88836.1 KQ434899 KZC11037.1 GBYB01010475 JAG80242.1 GBYB01011998 JAG81765.1 KZ288252 PBC31031.1 ADTU01012618 ADTU01012619 ADTU01012620 ADTU01012621 ADTU01012622 KQ982254 KYQ58732.1 KQ414617 KOC68131.1 QOIP01000002 RLU25619.1 HACA01031306 CDW48667.1 IACF01003718 LAB69329.1 KQ977830 KYM99406.1 KQ981082 KYN09659.1 GBYB01011999 JAG81766.1 GDIP01239226 GDIQ01174802 GDIQ01059790 JAI84175.1 JAK76923.1 GDIP01199822 JAJ23580.1 GDIP01168925 GDIQ01152317 JAJ54477.1 JAK99408.1 GDIP01052398 JAM51317.1 GDIP01186133 JAJ37269.1 GDIQ01172453 JAK79272.1 CRK99540.1 GDIP01109685 JAL94029.1 GDIP01197374 JAJ26028.1 KQ976424 KYM88257.1 GDIQ01057241 JAN37496.1 GDIQ01248331 GDIQ01090834 JAK03394.1 GDIP01057941 JAM45774.1 LRGB01000781 KZS15954.1 GDIQ01097072 JAL54654.1 GDIQ01219294 JAK32431.1 GDIP01239225 GDIQ01268974 GDIQ01237277 GDIQ01140514 GDIQ01130364 JAI84176.1 JAJ82750.1 GDIQ01108723 JAL43003.1 GDIQ01057240 JAN37497.1 GDIP01186132 JAJ37270.1 GDIQ01034708 JAN60029.1
Proteomes
UP000283053
UP000005204
UP000007151
UP000053268
UP000053240
UP000027135
+ More
UP000075840 UP000069272 UP000075920 UP000183832 UP000037069 UP000075883 UP000069940 UP000249989 UP000095301 UP000007266 UP000030765 UP000007801 UP000001819 UP000076407 UP000095300 UP000192221 UP000268350 UP000094527 UP000000803 UP000009046 UP000075886 UP000007062 UP000076408 UP000009192 UP000007798 UP000002282 UP000092460 UP000008792 UP000198287 UP000053097 UP000000311 UP000000305 UP000092445 UP000076502 UP000242457 UP000005205 UP000005203 UP000075809 UP000053825 UP000279307 UP000078542 UP000078492 UP000078540 UP000076858
UP000075840 UP000069272 UP000075920 UP000183832 UP000037069 UP000075883 UP000069940 UP000249989 UP000095301 UP000007266 UP000030765 UP000007801 UP000001819 UP000076407 UP000095300 UP000192221 UP000268350 UP000094527 UP000000803 UP000009046 UP000075886 UP000007062 UP000076408 UP000009192 UP000007798 UP000002282 UP000092460 UP000008792 UP000198287 UP000053097 UP000000311 UP000000305 UP000092445 UP000076502 UP000242457 UP000005205 UP000005203 UP000075809 UP000053825 UP000279307 UP000078542 UP000078492 UP000078540 UP000076858
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
A0A2H1V8C3
A0A3S2NLT4
H9JK88
A0A212FGA5
A0A194PQV4
A0A0N1IAT8
+ More
A0A194PSG0 A0A067QK27 A0A182I8N7 A0A182F9A9 A0A182W155 A0A1J1IH42 A0A0L0BPR8 A0A182MD52 A0A182GR80 A0A1I8M182 A0A139WLQ8 A0A084VEE9 B3LZK5 A0A0P9AP37 Q295P3 A0A0N8P1B3 A0A1Y9IZV1 A0A1I8QAT0 A0A0R3NJ28 A0A1W4VTB0 A0A3B0KH40 A0A1D2N0V7 A0A1W4VTZ4 A0A126GUS1 A0A126GUR3 Q9VHQ8 A0A0R3NP57 A0A0R3NQS8 A0A1W4VUN3 A0A1W4VGC4 B5X504 E0W270 A0A182QVU9 Q7QFT8 A0A182XX40 B4KD35 A0A1W4VGL5 B4N9A1 A0A0B4K609 A0A0R1E1N3 A0A0B4K642 A0A034WGA0 A0A1B0B5M4 A0A0A1XJT7 Q9VHR2 W8C0L2 W8B1Y8 B4LZM1 A0A226ETC0 A0A026W2F3 E2AQP1 F5HJC7 E9GS53 A0A0P4YQI9 A0A1B0AC74 A0A154PGV5 A0A0C9RPS0 A0A0C9QBZ9 A0A2A3EJ29 A0A158NE28 A0A088A2P4 A0A151XED5 A0A0L7RBK8 A0A3L8E0B7 A0A0K2VEP7 A0A2P2I5N8 A0A151IF28 A0A195DB68 A0A0C9R6P0 A0A0P4XRC3 A0A0P5BGU0 A0A0P5MWX0 A0A0P5YZD4 A0A0P5BDB4 A0A0P5LB48 A0A1J1IJ21 A0A0P5UTM1 A0A0P5AM18 A0A195BP15 A0A0P6EVR9 A0A0N8AS41 A0A0P5YHC0 A0A164Z592 A0A0P5S452 A0A0P5ICW1 A0A0P5EIR9 A0A0N8C6Z2 A0A0P6EVV5 A0A0P5BF28 A0A0N8EEH0
A0A194PSG0 A0A067QK27 A0A182I8N7 A0A182F9A9 A0A182W155 A0A1J1IH42 A0A0L0BPR8 A0A182MD52 A0A182GR80 A0A1I8M182 A0A139WLQ8 A0A084VEE9 B3LZK5 A0A0P9AP37 Q295P3 A0A0N8P1B3 A0A1Y9IZV1 A0A1I8QAT0 A0A0R3NJ28 A0A1W4VTB0 A0A3B0KH40 A0A1D2N0V7 A0A1W4VTZ4 A0A126GUS1 A0A126GUR3 Q9VHQ8 A0A0R3NP57 A0A0R3NQS8 A0A1W4VUN3 A0A1W4VGC4 B5X504 E0W270 A0A182QVU9 Q7QFT8 A0A182XX40 B4KD35 A0A1W4VGL5 B4N9A1 A0A0B4K609 A0A0R1E1N3 A0A0B4K642 A0A034WGA0 A0A1B0B5M4 A0A0A1XJT7 Q9VHR2 W8C0L2 W8B1Y8 B4LZM1 A0A226ETC0 A0A026W2F3 E2AQP1 F5HJC7 E9GS53 A0A0P4YQI9 A0A1B0AC74 A0A154PGV5 A0A0C9RPS0 A0A0C9QBZ9 A0A2A3EJ29 A0A158NE28 A0A088A2P4 A0A151XED5 A0A0L7RBK8 A0A3L8E0B7 A0A0K2VEP7 A0A2P2I5N8 A0A151IF28 A0A195DB68 A0A0C9R6P0 A0A0P4XRC3 A0A0P5BGU0 A0A0P5MWX0 A0A0P5YZD4 A0A0P5BDB4 A0A0P5LB48 A0A1J1IJ21 A0A0P5UTM1 A0A0P5AM18 A0A195BP15 A0A0P6EVR9 A0A0N8AS41 A0A0P5YHC0 A0A164Z592 A0A0P5S452 A0A0P5ICW1 A0A0P5EIR9 A0A0N8C6Z2 A0A0P6EVV5 A0A0P5BF28 A0A0N8EEH0
PDB
5LFU
E-value=8.25623e-14,
Score=189
Ontologies
GO
Topology
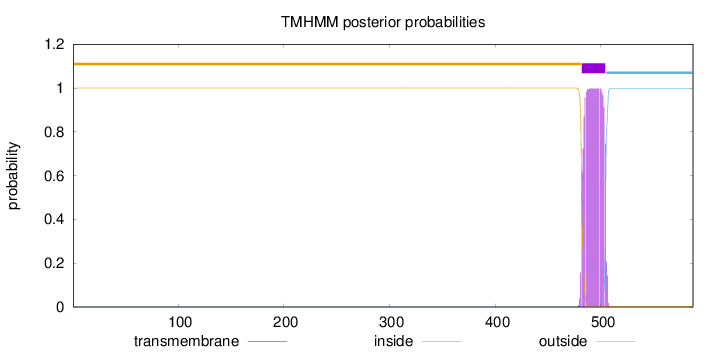
Length:
587
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.28398
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00003
outside
1 - 481
TMhelix
482 - 504
inside
505 - 587
Population Genetic Test Statistics
Pi
286.001524
Theta
163.320268
Tajima's D
2.624291
CLR
0.537563
CSRT
0.955102244887756
Interpretation
Uncertain
