Gene
KWMTBOMO04702
Annotation
PREDICTED:_G-protein_coupled_receptor_Mth2-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.94
Sequence
CDS
ATGTTGGTTAAAGTTTCGCTTCTATTTTCAATAACTTTGTCGTTTCGTTCAGTGGATTGTTTCGCTACCAAACTCAGTGAATTGGATATATGTAAAAATAAAATATGCATACCTAAATGCTGTGGTTTAGATAAATACGTGCACGGCAGAGATAGCAAGTGCGCCTACACCGATTTGAAGGATGCATTGAATTTTTCTAAAGTACCTGTCTACAAAGACGACTTAACAACGGTGGTAACAACGCTGACTGATCCGAGGTTCCATATGGTCGGGAATGTTTTCGAAGATGATGAATTCAAAAGTCTCGCCTACGACTTAAGTCGCACACGGATCTACAACTTTTTTTTATCTGAGACAGGTGTGCTCTACCTAGAGAAATCAAACTTTTACTTACGATGGACACCAAGGAACACTTCTAGATACTGCGTTGACTACAGAAAAGTGAAAAGCAAAAACGCCTCTGTGGCCTGGGAGGCACAATATCGTTTCTTTACTGTTATGGATCCTGTAGCTCAACCCGAAACCCAAACACTTTACAGAACTGCGCTGATGATTTCAAGTTTGTTTCTTTTAATTGTGCTTTTGGTCTACGCGATACTACCAGAGCTCAGGAACTTGGGTGGTTTGATTCTGATGGCATATGTAGGGAGTCTATTGTGCGCTTTCTTTTTGCTGTCTATCATACTGCTCGATATCAACTACAAAACAAACACTTGCATTGGAAGTACTATGGTGATATACTACAGTCTTATAGCAAGTTTTTGTTGGATGAACATCATGTCATACGACATTTGGTGGACATTTAGAGGATACGCTAAAGCAAGATCGATTCACCGTCGCGGTGAAATCTTCAAATTTCAAATGTACTGCTTGTATGGTTTTGGAGTACCTCTAGTCCTGACCATACTACTCGCTGTCATGCATTTCGTTGACTTTAGCGACCAGCCCTGGATAATCAAGCCACAGTTGGTACGCTACGGTTGCTTCATGGGAGGTCCCGAGAAAATGTTATACCTCTACGTACCGATGCTGATTCTAATCATGTGCAATTGGATGTTCTTCCTGATGACTGCTTTCAATGTGTGGCGGCTCAGTCGCGGTACGGCTGTCCTGGATTCGGCCGCTGCCGGAAACCCACACGCGCATCGCACTCAGAGACACAGGTTTATGGTGTATTTAAAGTTATCCATTGTGATGGGTATAAACTGGGTATTAGAAGTGGTTAGTTCTCAGCTCCCCGAGCTACAAATCTTCAAAGTTAGCGACCTATACAACGTCATGGTCGGCTTGTTCATATTCCTCATCTTCGTTTTCAAGAAGAAAATACTAAGGAAACTCATGAAAAAATTGCCGAACAAGAAGCGTTGGGCTCCGGCGTCGAAGAGCTACGAATCAGACAGCAGCACGTCAGACGGAAGCAGCCAAGAGACTTCCTTGCCTTTGGTTTGTTCAATCCCAAAGGGGCCTATCGAAAAGAGCACTTAA
Protein
MLVKVSLLFSITLSFRSVDCFATKLSELDICKNKICIPKCCGLDKYVHGRDSKCAYTDLKDALNFSKVPVYKDDLTTVVTTLTDPRFHMVGNVFEDDEFKSLAYDLSRTRIYNFFLSETGVLYLEKSNFYLRWTPRNTSRYCVDYRKVKSKNASVAWEAQYRFFTVMDPVAQPETQTLYRTALMISSLFLLIVLLVYAILPELRNLGGLILMAYVGSLLCAFFLLSIILLDINYKTNTCIGSTMVIYYSLIASFCWMNIMSYDIWWTFRGYAKARSIHRRGEIFKFQMYCLYGFGVPLVLTILLAVMHFVDFSDQPWIIKPQLVRYGCFMGGPEKMLYLYVPMLILIMCNWMFFLMTAFNVWRLSRGTAVLDSAAAGNPHAHRTQRHRFMVYLKLSIVMGINWVLEVVSSQLPELQIFKVSDLYNVMVGLFIFLIFVFKKKILRKLMKKLPNKKRWAPASKSYESDSSTSDGSSQETSLPLVCSIPKGPIEKST
Summary
Uniprot
Pubmed
EMBL
Proteomes
Interpro
IPR013083
Znf_RING/FYVE/PHD
+ More
IPR017981 GPCR_2-like
IPR016135 UBQ-conjugating_enzyme/RWD
IPR001841 Znf_RING
IPR006575 RWD-domain
IPR000477 RT_dom
IPR036691 Endo/exonu/phosph_ase_sf
IPR000832 GPCR_2_secretin-like
IPR005135 Endo/exonuclease/phosphatase
IPR036272 Methuselah_N_sf
IPR010596 Methuselah_N_dom
IPR017981 GPCR_2-like
IPR016135 UBQ-conjugating_enzyme/RWD
IPR001841 Znf_RING
IPR006575 RWD-domain
IPR000477 RT_dom
IPR036691 Endo/exonu/phosph_ase_sf
IPR000832 GPCR_2_secretin-like
IPR005135 Endo/exonuclease/phosphatase
IPR036272 Methuselah_N_sf
IPR010596 Methuselah_N_dom
Gene 3D
ProteinModelPortal
Ontologies
GO
Topology
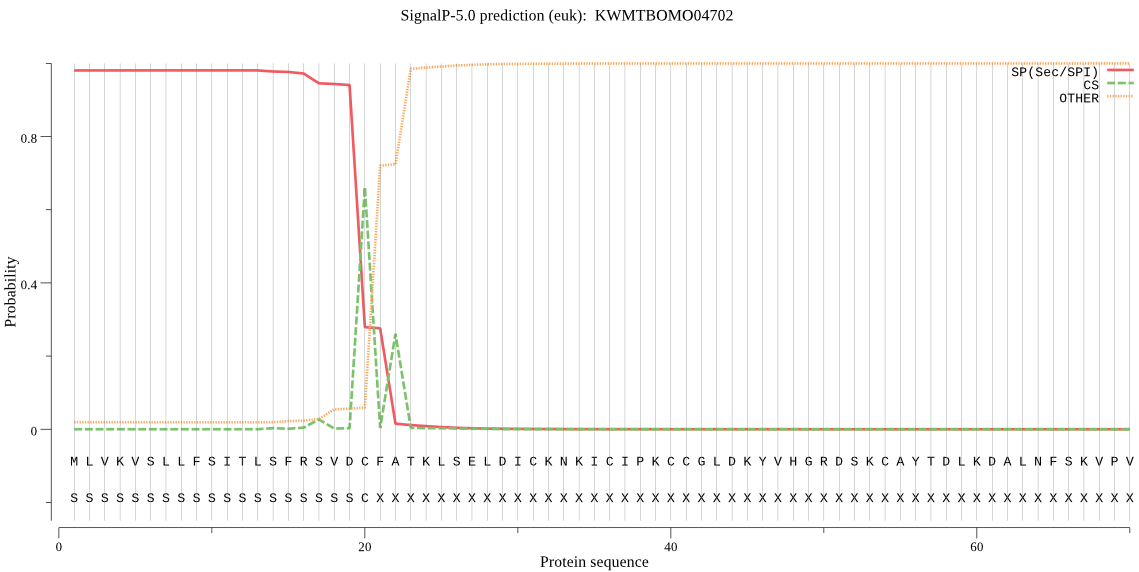
SignalP
Position: 1 - 20,
Likelihood: 0.980322
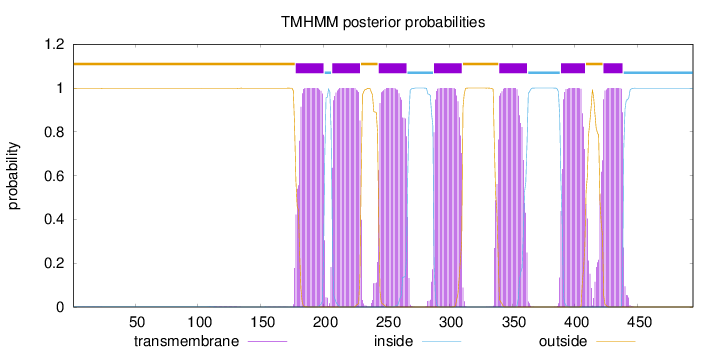
Length:
494
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
151.02044
Exp number, first 60 AAs:
0.00487
Total prob of N-in:
0.00261
outside
1 - 177
TMhelix
178 - 200
inside
201 - 206
TMhelix
207 - 229
outside
230 - 243
TMhelix
244 - 266
inside
267 - 287
TMhelix
288 - 310
outside
311 - 339
TMhelix
340 - 362
inside
363 - 388
TMhelix
389 - 408
outside
409 - 422
TMhelix
423 - 438
inside
439 - 494
Population Genetic Test Statistics
Pi
228.813987
Theta
156.018478
Tajima's D
1.536442
CLR
0.348086
CSRT
0.794410279486026
Interpretation
Uncertain
