Gene
KWMTBOMO04699
Pre Gene Modal
BGIBMGA009811
Annotation
PREDICTED:_E3_ubiquitin-protein_ligase_RNF25_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.073
Sequence
CDS
ATGTCGTCCTCCGGCGATGAAAGAATCACGGACGAGGTGCAAGCATTAGAAGCAATTCTACTGAAAGATGTGGTAGTAAAACGCGTTGGAGATGTCCCAAAATACATAATTGAAACGATCATCCATCCGTTGACGGGGGATAATATCGATCAGCAATATGTATGTGTGACTCTGGAGGTGGGGTTGTCTTCTGGCTATCCAGACACCAGCCCAGAAGTAGCGCTCAAAAACCCTAGAGGTCTAGATGATATGATTTTAGACACTATCAACACACATATTCAAGACAAATTGAAAAATAATTTGGGACAACCAATAGTGTTTGAACTTATTGGAATTGTACGTGATCATCTGACTAAGAGCAATCTTCCTCGTGGCCAGTGTGTCATATGCCTATATGATTTTGTCGATGGTGATTTGTTTATAAAGACGCAATGCTATCATTACTTCCACAATCATTGCCTTGCAAACCATTTCATCGCTGGCAAGAAATATTATCACGAGGAACTTGAGAAACTACCTACTTGGCAACAGATGCAAGTGCCACCTTACCAGCAAACATGCCCGGTGTGTCGTTGCACGGTATCATGTGACGTGGACACTTTAAAGGAGGCTCCCCCACCAAAGAGCTCCCTGTCGGCACCACCGTTCCGCTTGACAGCTGAACTAAAAGAACTCCAAATGAAGATGGCCGAACTGTTAGCTGTGCAAGTTGCTAAGGGCGGGGTCGTCGGTATGGGCGACAATGGGCCCCCCCTGCTGACCATCACAACCACACCCACTACTGACAACGTAAATATATAA
Protein
MSSSGDERITDEVQALEAILLKDVVVKRVGDVPKYIIETIIHPLTGDNIDQQYVCVTLEVGLSSGYPDTSPEVALKNPRGLDDMILDTINTHIQDKLKNNLGQPIVFELIGIVRDHLTKSNLPRGQCVICLYDFVDGDLFIKTQCYHYFHNHCLANHFIAGKKYYHEELEKLPTWQQMQVPPYQQTCPVCRCTVSCDVDTLKEAPPPKSSLSAPPFRLTAELKELQMKMAELLAVQVAKGGVVGMGDNGPPLLTITTTPTTDNVNI
Summary
Uniprot
A0A3S2PGP9
A0A194QUF4
A0A194PX16
A0A2A4IUG3
A0A212FA89
A0A2H1V0J5
+ More
E2BKN7 A0A2A3EHP2 V9IHQ8 A0A088ANP8 A0A3L8E001 A0A026X2H9 A0A0L7R149 A0A0J7KVX0 E2AFI1 A0A195CTI8 A0A151XG69 A0A310SXS8 A0A182JIR8 B0WTE6 A0A1I8MGJ8 A0A182GZS7 A0A1S4FTL6 Q0IEB3 A0A3B0J5R2 A0A182TRH5 A0A182UMM5 A0A182WZM3 B4N5W9 Q7Q096 A0A182HQ69 A0A182U045 A0A182QC07 A0A084WHJ1 A0A182Y1N9 A0A182MYN9 A0A182KSI0 A0A154PB13 B3MHF5 A0A067RR42 A0A182K3C1 J3JVA2 A0A232F5M5 A0A0L0BSR7 A0A1I8NX45 K7IV54 A0A1B0BLN5 B4GGS3 A0A1A9Y660 B5E0Y1 A0A1A9VXI0 A0A336KUQ1 A0A1W4V4A2 A0A1B0A7E3 A0A1B0FKY3 A0A0N0BEB7 A0A182LXV0 A0A182WG66 B4HQY2 W5JGG0 A0A182RTJ5 B4QEV3 A0A1A9W9U0 A8WHE3 A1Z9I6 A0A182FW62 D0IQC3 A0A151JPX0 A0A2M4AWX2 B3NRF9 A0A195BZ22 A0A0J9RD95 A0A195F0D9 E9IK81 B4P454 D7EJZ3 A0A2M4AWF3 A0A182P6I6 A0A2J7PU62 B4KTB2 A0A224XTY9 A0A0K8VBF5 A0A034WHM0 A0A0K8U6C7 A0A0A1WMP1 A0A0M3QUS6 A0A1B6CUG0 A0A2J7PU36 Q7K199 A0A0A9YWV5 B4LJF6 B4JW25 A0A1B6GI42 W8B4X8 A0A1B6IK36 A0A182SXY6 A0A2R7W482 A0A2P8YR14 A0A1B6LJV0
E2BKN7 A0A2A3EHP2 V9IHQ8 A0A088ANP8 A0A3L8E001 A0A026X2H9 A0A0L7R149 A0A0J7KVX0 E2AFI1 A0A195CTI8 A0A151XG69 A0A310SXS8 A0A182JIR8 B0WTE6 A0A1I8MGJ8 A0A182GZS7 A0A1S4FTL6 Q0IEB3 A0A3B0J5R2 A0A182TRH5 A0A182UMM5 A0A182WZM3 B4N5W9 Q7Q096 A0A182HQ69 A0A182U045 A0A182QC07 A0A084WHJ1 A0A182Y1N9 A0A182MYN9 A0A182KSI0 A0A154PB13 B3MHF5 A0A067RR42 A0A182K3C1 J3JVA2 A0A232F5M5 A0A0L0BSR7 A0A1I8NX45 K7IV54 A0A1B0BLN5 B4GGS3 A0A1A9Y660 B5E0Y1 A0A1A9VXI0 A0A336KUQ1 A0A1W4V4A2 A0A1B0A7E3 A0A1B0FKY3 A0A0N0BEB7 A0A182LXV0 A0A182WG66 B4HQY2 W5JGG0 A0A182RTJ5 B4QEV3 A0A1A9W9U0 A8WHE3 A1Z9I6 A0A182FW62 D0IQC3 A0A151JPX0 A0A2M4AWX2 B3NRF9 A0A195BZ22 A0A0J9RD95 A0A195F0D9 E9IK81 B4P454 D7EJZ3 A0A2M4AWF3 A0A182P6I6 A0A2J7PU62 B4KTB2 A0A224XTY9 A0A0K8VBF5 A0A034WHM0 A0A0K8U6C7 A0A0A1WMP1 A0A0M3QUS6 A0A1B6CUG0 A0A2J7PU36 Q7K199 A0A0A9YWV5 B4LJF6 B4JW25 A0A1B6GI42 W8B4X8 A0A1B6IK36 A0A182SXY6 A0A2R7W482 A0A2P8YR14 A0A1B6LJV0
Pubmed
26354079
22118469
20798317
30249741
24508170
25315136
+ More
26483478 17510324 17994087 12364791 14747013 17210077 24438588 25244985 20966253 24845553 22516182 23537049 28648823 26108605 20075255 15632085 20920257 23761445 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 17550304 18362917 19820115 25348373 25830018 25401762 26823975 24495485 29403074
26483478 17510324 17994087 12364791 14747013 17210077 24438588 25244985 20966253 24845553 22516182 23537049 28648823 26108605 20075255 15632085 20920257 23761445 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 17550304 18362917 19820115 25348373 25830018 25401762 26823975 24495485 29403074
EMBL
RSAL01000041
RVE50932.1
KQ461154
KPJ08610.1
KQ459595
KPI95685.1
+ More
NWSH01006493 PCG63395.1 AGBW02009506 OWR50656.1 ODYU01000121 SOQ34357.1 GL448819 EFN83735.1 KZ288254 PBC30792.1 JR048385 AEY60648.1 QOIP01000002 RLU25882.1 KK107026 EZA62201.1 KQ414668 KOC64573.1 LBMM01002582 KMQ94627.1 GL439108 EFN67874.1 KQ977304 KYN03807.1 KQ982174 KYQ59347.1 KQ759767 OAD62959.1 DS232085 EDS34384.1 JXUM01002155 JXUM01002156 JXUM01002157 CH477764 EAT36498.1 OUUW01000001 SPP75022.1 CH964154 EDW79758.1 AAAB01008986 EAA00366.4 APCN01003297 AXCN02001035 ATLV01023841 KE525347 KFB49685.1 KQ434850 KZC08574.1 CH902619 EDV37955.1 KK852473 KDR23110.1 APGK01055664 BT127169 KB741269 KB632330 AEE62131.1 ENN71428.1 ERL92600.1 NNAY01000950 OXU25763.1 JRES01001421 KNC23061.1 AAZX01007538 JXJN01014763 JXJN01016503 CH479183 EDW35693.1 CM000071 EDY69062.1 UFQS01000845 UFQT01000845 SSX07245.1 SSX27588.1 CCAG010009481 KQ435833 KOX71654.1 AXCM01001617 CH480816 EDW47779.1 ADMH02001285 ETN63176.1 CM000362 CM002911 EDX06980.1 KMY93588.1 BT031089 ABX00711.1 AE013599 AAF58321.2 AAZ52807.2 BT100161 ACY07066.1 KQ978744 KYN28698.1 GGFK01011975 MBW45296.1 CH954179 EDV56111.1 KQ976394 KYM93176.1 KMY93589.1 KQ981880 KYN33943.1 GL763924 EFZ19029.1 CM000158 EDW90564.2 KQ972213 EFA12922.2 GGFK01011802 MBW45123.1 NEVH01021208 PNF19856.1 CH933808 EDW10624.1 GFTR01004444 JAW11982.1 GDHF01016454 JAI35860.1 GAKP01005714 JAC53238.1 GDHF01030102 JAI22212.1 GBXI01014599 JAC99692.1 CP012524 ALC41179.1 GEDC01020435 JAS16863.1 PNF19855.1 AY069251 AAL39396.1 GBHO01034308 GBHO01006930 GBRD01009174 GDHC01004518 GDHC01000143 JAG09296.1 JAG36674.1 JAG56647.1 JAQ14111.1 JAQ18486.1 CH940648 EDW60536.1 CH916375 EDV98163.1 GECZ01007665 JAS62104.1 GAMC01014417 JAB92138.1 GECU01020438 JAS87268.1 KK854313 PTY14527.1 PYGN01000420 PSN46697.1 GEBQ01016012 JAT23965.1
NWSH01006493 PCG63395.1 AGBW02009506 OWR50656.1 ODYU01000121 SOQ34357.1 GL448819 EFN83735.1 KZ288254 PBC30792.1 JR048385 AEY60648.1 QOIP01000002 RLU25882.1 KK107026 EZA62201.1 KQ414668 KOC64573.1 LBMM01002582 KMQ94627.1 GL439108 EFN67874.1 KQ977304 KYN03807.1 KQ982174 KYQ59347.1 KQ759767 OAD62959.1 DS232085 EDS34384.1 JXUM01002155 JXUM01002156 JXUM01002157 CH477764 EAT36498.1 OUUW01000001 SPP75022.1 CH964154 EDW79758.1 AAAB01008986 EAA00366.4 APCN01003297 AXCN02001035 ATLV01023841 KE525347 KFB49685.1 KQ434850 KZC08574.1 CH902619 EDV37955.1 KK852473 KDR23110.1 APGK01055664 BT127169 KB741269 KB632330 AEE62131.1 ENN71428.1 ERL92600.1 NNAY01000950 OXU25763.1 JRES01001421 KNC23061.1 AAZX01007538 JXJN01014763 JXJN01016503 CH479183 EDW35693.1 CM000071 EDY69062.1 UFQS01000845 UFQT01000845 SSX07245.1 SSX27588.1 CCAG010009481 KQ435833 KOX71654.1 AXCM01001617 CH480816 EDW47779.1 ADMH02001285 ETN63176.1 CM000362 CM002911 EDX06980.1 KMY93588.1 BT031089 ABX00711.1 AE013599 AAF58321.2 AAZ52807.2 BT100161 ACY07066.1 KQ978744 KYN28698.1 GGFK01011975 MBW45296.1 CH954179 EDV56111.1 KQ976394 KYM93176.1 KMY93589.1 KQ981880 KYN33943.1 GL763924 EFZ19029.1 CM000158 EDW90564.2 KQ972213 EFA12922.2 GGFK01011802 MBW45123.1 NEVH01021208 PNF19856.1 CH933808 EDW10624.1 GFTR01004444 JAW11982.1 GDHF01016454 JAI35860.1 GAKP01005714 JAC53238.1 GDHF01030102 JAI22212.1 GBXI01014599 JAC99692.1 CP012524 ALC41179.1 GEDC01020435 JAS16863.1 PNF19855.1 AY069251 AAL39396.1 GBHO01034308 GBHO01006930 GBRD01009174 GDHC01004518 GDHC01000143 JAG09296.1 JAG36674.1 JAG56647.1 JAQ14111.1 JAQ18486.1 CH940648 EDW60536.1 CH916375 EDV98163.1 GECZ01007665 JAS62104.1 GAMC01014417 JAB92138.1 GECU01020438 JAS87268.1 KK854313 PTY14527.1 PYGN01000420 PSN46697.1 GEBQ01016012 JAT23965.1
Proteomes
UP000283053
UP000053240
UP000053268
UP000218220
UP000007151
UP000008237
+ More
UP000242457 UP000005203 UP000279307 UP000053097 UP000053825 UP000036403 UP000000311 UP000078542 UP000075809 UP000075880 UP000002320 UP000095301 UP000069940 UP000008820 UP000268350 UP000075902 UP000075903 UP000076407 UP000007798 UP000007062 UP000075840 UP000075886 UP000030765 UP000076408 UP000075884 UP000075882 UP000076502 UP000007801 UP000027135 UP000075881 UP000019118 UP000030742 UP000215335 UP000037069 UP000095300 UP000002358 UP000092460 UP000008744 UP000092443 UP000001819 UP000078200 UP000192221 UP000092445 UP000092444 UP000053105 UP000075883 UP000075920 UP000001292 UP000000673 UP000075900 UP000000304 UP000091820 UP000000803 UP000069272 UP000078492 UP000008711 UP000078540 UP000078541 UP000002282 UP000007266 UP000075885 UP000235965 UP000009192 UP000092553 UP000008792 UP000001070 UP000075901 UP000245037
UP000242457 UP000005203 UP000279307 UP000053097 UP000053825 UP000036403 UP000000311 UP000078542 UP000075809 UP000075880 UP000002320 UP000095301 UP000069940 UP000008820 UP000268350 UP000075902 UP000075903 UP000076407 UP000007798 UP000007062 UP000075840 UP000075886 UP000030765 UP000076408 UP000075884 UP000075882 UP000076502 UP000007801 UP000027135 UP000075881 UP000019118 UP000030742 UP000215335 UP000037069 UP000095300 UP000002358 UP000092460 UP000008744 UP000092443 UP000001819 UP000078200 UP000192221 UP000092445 UP000092444 UP000053105 UP000075883 UP000075920 UP000001292 UP000000673 UP000075900 UP000000304 UP000091820 UP000000803 UP000069272 UP000078492 UP000008711 UP000078540 UP000078541 UP000002282 UP000007266 UP000075885 UP000235965 UP000009192 UP000092553 UP000008792 UP000001070 UP000075901 UP000245037
Interpro
SUPFAM
SSF54495
SSF54495
Gene 3D
ProteinModelPortal
A0A3S2PGP9
A0A194QUF4
A0A194PX16
A0A2A4IUG3
A0A212FA89
A0A2H1V0J5
+ More
E2BKN7 A0A2A3EHP2 V9IHQ8 A0A088ANP8 A0A3L8E001 A0A026X2H9 A0A0L7R149 A0A0J7KVX0 E2AFI1 A0A195CTI8 A0A151XG69 A0A310SXS8 A0A182JIR8 B0WTE6 A0A1I8MGJ8 A0A182GZS7 A0A1S4FTL6 Q0IEB3 A0A3B0J5R2 A0A182TRH5 A0A182UMM5 A0A182WZM3 B4N5W9 Q7Q096 A0A182HQ69 A0A182U045 A0A182QC07 A0A084WHJ1 A0A182Y1N9 A0A182MYN9 A0A182KSI0 A0A154PB13 B3MHF5 A0A067RR42 A0A182K3C1 J3JVA2 A0A232F5M5 A0A0L0BSR7 A0A1I8NX45 K7IV54 A0A1B0BLN5 B4GGS3 A0A1A9Y660 B5E0Y1 A0A1A9VXI0 A0A336KUQ1 A0A1W4V4A2 A0A1B0A7E3 A0A1B0FKY3 A0A0N0BEB7 A0A182LXV0 A0A182WG66 B4HQY2 W5JGG0 A0A182RTJ5 B4QEV3 A0A1A9W9U0 A8WHE3 A1Z9I6 A0A182FW62 D0IQC3 A0A151JPX0 A0A2M4AWX2 B3NRF9 A0A195BZ22 A0A0J9RD95 A0A195F0D9 E9IK81 B4P454 D7EJZ3 A0A2M4AWF3 A0A182P6I6 A0A2J7PU62 B4KTB2 A0A224XTY9 A0A0K8VBF5 A0A034WHM0 A0A0K8U6C7 A0A0A1WMP1 A0A0M3QUS6 A0A1B6CUG0 A0A2J7PU36 Q7K199 A0A0A9YWV5 B4LJF6 B4JW25 A0A1B6GI42 W8B4X8 A0A1B6IK36 A0A182SXY6 A0A2R7W482 A0A2P8YR14 A0A1B6LJV0
E2BKN7 A0A2A3EHP2 V9IHQ8 A0A088ANP8 A0A3L8E001 A0A026X2H9 A0A0L7R149 A0A0J7KVX0 E2AFI1 A0A195CTI8 A0A151XG69 A0A310SXS8 A0A182JIR8 B0WTE6 A0A1I8MGJ8 A0A182GZS7 A0A1S4FTL6 Q0IEB3 A0A3B0J5R2 A0A182TRH5 A0A182UMM5 A0A182WZM3 B4N5W9 Q7Q096 A0A182HQ69 A0A182U045 A0A182QC07 A0A084WHJ1 A0A182Y1N9 A0A182MYN9 A0A182KSI0 A0A154PB13 B3MHF5 A0A067RR42 A0A182K3C1 J3JVA2 A0A232F5M5 A0A0L0BSR7 A0A1I8NX45 K7IV54 A0A1B0BLN5 B4GGS3 A0A1A9Y660 B5E0Y1 A0A1A9VXI0 A0A336KUQ1 A0A1W4V4A2 A0A1B0A7E3 A0A1B0FKY3 A0A0N0BEB7 A0A182LXV0 A0A182WG66 B4HQY2 W5JGG0 A0A182RTJ5 B4QEV3 A0A1A9W9U0 A8WHE3 A1Z9I6 A0A182FW62 D0IQC3 A0A151JPX0 A0A2M4AWX2 B3NRF9 A0A195BZ22 A0A0J9RD95 A0A195F0D9 E9IK81 B4P454 D7EJZ3 A0A2M4AWF3 A0A182P6I6 A0A2J7PU62 B4KTB2 A0A224XTY9 A0A0K8VBF5 A0A034WHM0 A0A0K8U6C7 A0A0A1WMP1 A0A0M3QUS6 A0A1B6CUG0 A0A2J7PU36 Q7K199 A0A0A9YWV5 B4LJF6 B4JW25 A0A1B6GI42 W8B4X8 A0A1B6IK36 A0A182SXY6 A0A2R7W482 A0A2P8YR14 A0A1B6LJV0
PDB
2DMF
E-value=8.81247e-16,
Score=202
Ontologies
GO
PANTHER
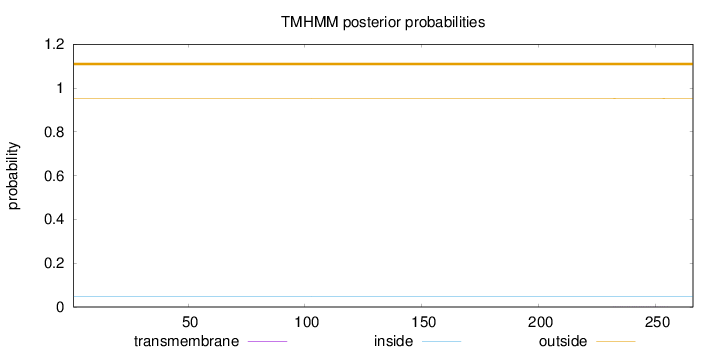
Topology
Length:
266
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00649
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04829
outside
1 - 266
Population Genetic Test Statistics
Pi
181.219299
Theta
178.902528
Tajima's D
-0.429042
CLR
11.7967
CSRT
0.251287435628219
Interpretation
Uncertain
