Gene
KWMTBOMO04698
Annotation
PREDICTED:_switch-associated_protein_70-like_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.664 Nuclear Reliability : 2.153
Sequence
CDS
ATGGTACTAACCGCAAACATAGGCACCCTACTTGACCTGTACGGCGTGGAAAAAGGTCTGGACCACTTCAGGTCATCCCAGGAGTTGACATTCGACGAGTTCAAATACTATCTCCAGCACGAAGTCTTCAGTTCCCTGCCGAAGACGGTAACTCTGCCTATCATCAGGGAATATGAGAAGAAAATAAGCGAGGTGTGCTGGCTGGTCTGCCGTAAGCGTCTCCTCTTTAGGGACAAGTCGGTTTTCGGAGACGACTCCGTTTTCAAGCTCTTCAGGATATTCTGTCTTCTCGCTGACTTGGTTCAAGACGCTGACGACACCAATCGATATGTGAGTAATGCTGAAAGAAAAAAGACCACCGAGAATCACTGA
Protein
MVLTANIGTLLDLYGVEKGLDHFRSSQELTFDEFKYYLQHEVFSSLPKTVTLPIIREYEKKISEVCWLVCRKRLLFRDKSVFGDDSVFKLFRIFCLLADLVQDADDTNRYVSNAERKKTTENH
Summary
Similarity
Belongs to the eukaryotic ribosomal protein P1/P2 family.
Uniprot
A0A2A4JEH6
A0A2A4ITN1
A0A2A4JE48
H9JJW1
A0A194QTH0
A0A194PRV1
+ More
A0A2H1V0S1 A0A212FA78 U4UTB7 N6TDY9 A0A1Y1M8C8 D6WYK8 A0A2P8YR10 A0A1B6IAW4 A0A1B6CFM6 A0A1B6L3D9 A0A151WSS5 A0A2J7Q8G9 A0A195CPX0 A0A195BXH2 A0A158NWP1 A0A195FPR1 E2BKC9 F4W3T0 A0A1B0DP54 E1ZZE2 A0A067RR30 A0A182RTJ3 A0A151JQV3 A0A026W3D5 A0A182P6I5 A0A3L8DBV7 E0VU53 A0A0J7KSS4 A0A182MHQ8 A0A1L8DV35 A0A1L8DV31 A0A182WG67 A0A1B0CIX4 K7IRU2 A0A182FW63 A0A3Q0JCB3 A0A084WHJ3 A0A2S2PE55 A0A232FHF5 A0A182USD3 A0A182TM66 A0A182Y1P0 A0A182KSH9 A0A182WZM2 A0NGI8 A0A182HQ68 W5JJU5 A0A182S991 A0A182QB47 A0A2R7VZ36 T1HMF8 A0A182IYD8 A0A182MYN8 E9ID97 A0A023F3R7 J9KB48 Q0IEB4 A0A1S4FTH6 A0A182GZS8 A0A336MC90 B0WTE5 A0A2S2R1M2 A0A1W4WIZ0 T1J5D9 A0A1W4WTU3 B7PKR8 A0A131YXS5 A0A224Y904 V5HNP7 A0A2R5LLS7 A0A1D2N3V1 A0A0K2U676 A0A1Z5KV15 A0A226F844 A0A1S3HX72 A0A1S3HVT9 A0A3R7QIS3 A0A0L8GL05 W4XT47
A0A2H1V0S1 A0A212FA78 U4UTB7 N6TDY9 A0A1Y1M8C8 D6WYK8 A0A2P8YR10 A0A1B6IAW4 A0A1B6CFM6 A0A1B6L3D9 A0A151WSS5 A0A2J7Q8G9 A0A195CPX0 A0A195BXH2 A0A158NWP1 A0A195FPR1 E2BKC9 F4W3T0 A0A1B0DP54 E1ZZE2 A0A067RR30 A0A182RTJ3 A0A151JQV3 A0A026W3D5 A0A182P6I5 A0A3L8DBV7 E0VU53 A0A0J7KSS4 A0A182MHQ8 A0A1L8DV35 A0A1L8DV31 A0A182WG67 A0A1B0CIX4 K7IRU2 A0A182FW63 A0A3Q0JCB3 A0A084WHJ3 A0A2S2PE55 A0A232FHF5 A0A182USD3 A0A182TM66 A0A182Y1P0 A0A182KSH9 A0A182WZM2 A0NGI8 A0A182HQ68 W5JJU5 A0A182S991 A0A182QB47 A0A2R7VZ36 T1HMF8 A0A182IYD8 A0A182MYN8 E9ID97 A0A023F3R7 J9KB48 Q0IEB4 A0A1S4FTH6 A0A182GZS8 A0A336MC90 B0WTE5 A0A2S2R1M2 A0A1W4WIZ0 T1J5D9 A0A1W4WTU3 B7PKR8 A0A131YXS5 A0A224Y904 V5HNP7 A0A2R5LLS7 A0A1D2N3V1 A0A0K2U676 A0A1Z5KV15 A0A226F844 A0A1S3HX72 A0A1S3HVT9 A0A3R7QIS3 A0A0L8GL05 W4XT47
Pubmed
19121390
26354079
22118469
23537049
28004739
18362917
+ More
19820115 29403074 21347285 20798317 21719571 24845553 24508170 30249741 20566863 20075255 24438588 28648823 25244985 20966253 12364791 14747013 17210077 20920257 23761445 21282665 25474469 17510324 26483478 26830274 28797301 25765539 27289101 28528879
19820115 29403074 21347285 20798317 21719571 24845553 24508170 30249741 20566863 20075255 24438588 28648823 25244985 20966253 12364791 14747013 17210077 20920257 23761445 21282665 25474469 17510324 26483478 26830274 28797301 25765539 27289101 28528879
EMBL
NWSH01001798
PCG70108.1
NWSH01007111
PCG63081.1
PCG70109.1
BABH01040642
+ More
BABH01040643 BABH01040644 KQ461154 KPJ08609.1 KQ459595 KPI95683.1 ODYU01000121 SOQ34356.1 AGBW02009506 OWR50655.1 KB632356 ERL93421.1 APGK01041556 KB740994 ENN75968.1 GEZM01037769 JAV81944.1 KQ971362 EFA09002.1 PYGN01000420 PSN46675.1 GECU01023648 JAS84058.1 GEDC01025087 JAS12211.1 GEBQ01021725 JAT18252.1 KQ982769 KYQ50930.1 NEVH01016952 PNF24878.1 KQ977444 KYN02756.1 KQ976401 KYM92648.1 ADTU01028285 KQ981335 KYN42580.1 GL448783 EFN83853.1 GL887413 EGI71143.1 AJVK01018087 AJVK01018088 GL435343 EFN73434.1 KK852473 KDR23100.1 KQ978622 KYN29737.1 KK107455 EZA50538.1 QOIP01000010 RLU17388.1 DS235780 EEB16909.1 LBMM01003642 KMQ93284.1 AXCM01002364 GFDF01003805 JAV10279.1 GFDF01003804 JAV10280.1 AJWK01013857 ATLV01023841 ATLV01023842 KE525347 KFB49687.1 GGMR01015053 MBY27672.1 NNAY01000187 OXU30174.1 AAAB01008986 EAU75769.2 APCN01003297 ADMH02001285 ETN63175.1 AXCN02001035 AXCN02001036 KK854184 PTY12753.1 ACPB03023598 GL762454 EFZ21431.1 GBBI01002662 JAC16050.1 ABLF02030115 CH477764 EAT36497.1 JXUM01002158 JXUM01002159 JXUM01002160 UFQT01000640 SSX26007.1 DS232085 EDS34383.1 GGMS01014467 MBY83670.1 JH431859 ABJB010557738 ABJB010915283 ABJB010970881 DS735295 EEC07190.1 GEDV01005826 JAP82731.1 GFPF01000903 MAA12049.1 GANP01009085 JAB75383.1 GGLE01006347 MBY10473.1 LJIJ01000244 ODM99948.1 HACA01016056 CDW33417.1 GFJQ02008041 JAV98928.1 LNIX01000001 OXA65026.1 QCYY01001139 ROT80237.1 KQ421395 KOF77549.1 AAGJ04164213 AAGJ04164214 AAGJ04164215 AAGJ04164216 AAGJ04164217 AAGJ04164218
BABH01040643 BABH01040644 KQ461154 KPJ08609.1 KQ459595 KPI95683.1 ODYU01000121 SOQ34356.1 AGBW02009506 OWR50655.1 KB632356 ERL93421.1 APGK01041556 KB740994 ENN75968.1 GEZM01037769 JAV81944.1 KQ971362 EFA09002.1 PYGN01000420 PSN46675.1 GECU01023648 JAS84058.1 GEDC01025087 JAS12211.1 GEBQ01021725 JAT18252.1 KQ982769 KYQ50930.1 NEVH01016952 PNF24878.1 KQ977444 KYN02756.1 KQ976401 KYM92648.1 ADTU01028285 KQ981335 KYN42580.1 GL448783 EFN83853.1 GL887413 EGI71143.1 AJVK01018087 AJVK01018088 GL435343 EFN73434.1 KK852473 KDR23100.1 KQ978622 KYN29737.1 KK107455 EZA50538.1 QOIP01000010 RLU17388.1 DS235780 EEB16909.1 LBMM01003642 KMQ93284.1 AXCM01002364 GFDF01003805 JAV10279.1 GFDF01003804 JAV10280.1 AJWK01013857 ATLV01023841 ATLV01023842 KE525347 KFB49687.1 GGMR01015053 MBY27672.1 NNAY01000187 OXU30174.1 AAAB01008986 EAU75769.2 APCN01003297 ADMH02001285 ETN63175.1 AXCN02001035 AXCN02001036 KK854184 PTY12753.1 ACPB03023598 GL762454 EFZ21431.1 GBBI01002662 JAC16050.1 ABLF02030115 CH477764 EAT36497.1 JXUM01002158 JXUM01002159 JXUM01002160 UFQT01000640 SSX26007.1 DS232085 EDS34383.1 GGMS01014467 MBY83670.1 JH431859 ABJB010557738 ABJB010915283 ABJB010970881 DS735295 EEC07190.1 GEDV01005826 JAP82731.1 GFPF01000903 MAA12049.1 GANP01009085 JAB75383.1 GGLE01006347 MBY10473.1 LJIJ01000244 ODM99948.1 HACA01016056 CDW33417.1 GFJQ02008041 JAV98928.1 LNIX01000001 OXA65026.1 QCYY01001139 ROT80237.1 KQ421395 KOF77549.1 AAGJ04164213 AAGJ04164214 AAGJ04164215 AAGJ04164216 AAGJ04164217 AAGJ04164218
Proteomes
UP000218220
UP000005204
UP000053240
UP000053268
UP000007151
UP000030742
+ More
UP000019118 UP000007266 UP000245037 UP000075809 UP000235965 UP000078542 UP000078540 UP000005205 UP000078541 UP000008237 UP000007755 UP000092462 UP000000311 UP000027135 UP000075900 UP000078492 UP000053097 UP000075885 UP000279307 UP000009046 UP000036403 UP000075883 UP000075920 UP000092461 UP000002358 UP000069272 UP000079169 UP000030765 UP000215335 UP000075903 UP000075902 UP000076408 UP000075882 UP000076407 UP000007062 UP000075840 UP000000673 UP000075901 UP000075886 UP000015103 UP000075880 UP000075884 UP000007819 UP000008820 UP000069940 UP000002320 UP000192223 UP000001555 UP000094527 UP000198287 UP000085678 UP000283509 UP000053454 UP000007110
UP000019118 UP000007266 UP000245037 UP000075809 UP000235965 UP000078542 UP000078540 UP000005205 UP000078541 UP000008237 UP000007755 UP000092462 UP000000311 UP000027135 UP000075900 UP000078492 UP000053097 UP000075885 UP000279307 UP000009046 UP000036403 UP000075883 UP000075920 UP000092461 UP000002358 UP000069272 UP000079169 UP000030765 UP000215335 UP000075903 UP000075902 UP000076408 UP000075882 UP000076407 UP000007062 UP000075840 UP000000673 UP000075901 UP000075886 UP000015103 UP000075880 UP000075884 UP000007819 UP000008820 UP000069940 UP000002320 UP000192223 UP000001555 UP000094527 UP000198287 UP000085678 UP000283509 UP000053454 UP000007110
PRIDE
Pfam
Interpro
IPR001849
PH_domain
+ More
IPR011993 PH-like_dom_sf
IPR020892 Cyclophilin-type_PPIase_CS
IPR029000 Cyclophilin-like_dom_sf
IPR003613 Ubox_domain
IPR002130 Cyclophilin-type_PPIase_dom
IPR013083 Znf_RING/FYVE/PHD
IPR039505 DRC1/2_N
IPR017441 Protein_kinase_ATP_BS
IPR000306 Znf_FYVE
IPR011009 Kinase-like_dom_sf
IPR029021 Prot-tyrosine_phosphatase-like
IPR038716 P1/P2_N_sf
IPR011011 Znf_FYVE_PHD
IPR003595 Tyr_Pase_cat
IPR000719 Prot_kinase_dom
IPR016130 Tyr_Pase_AS
IPR017455 Znf_FYVE-rel
IPR027534 Ribosomal_L12
IPR010569 Myotubularin-like_Pase_dom
IPR011992 EF-hand-dom_pair
IPR011993 PH-like_dom_sf
IPR020892 Cyclophilin-type_PPIase_CS
IPR029000 Cyclophilin-like_dom_sf
IPR003613 Ubox_domain
IPR002130 Cyclophilin-type_PPIase_dom
IPR013083 Znf_RING/FYVE/PHD
IPR039505 DRC1/2_N
IPR017441 Protein_kinase_ATP_BS
IPR000306 Znf_FYVE
IPR011009 Kinase-like_dom_sf
IPR029021 Prot-tyrosine_phosphatase-like
IPR038716 P1/P2_N_sf
IPR011011 Znf_FYVE_PHD
IPR003595 Tyr_Pase_cat
IPR000719 Prot_kinase_dom
IPR016130 Tyr_Pase_AS
IPR017455 Znf_FYVE-rel
IPR027534 Ribosomal_L12
IPR010569 Myotubularin-like_Pase_dom
IPR011992 EF-hand-dom_pair
SUPFAM
Gene 3D
ProteinModelPortal
A0A2A4JEH6
A0A2A4ITN1
A0A2A4JE48
H9JJW1
A0A194QTH0
A0A194PRV1
+ More
A0A2H1V0S1 A0A212FA78 U4UTB7 N6TDY9 A0A1Y1M8C8 D6WYK8 A0A2P8YR10 A0A1B6IAW4 A0A1B6CFM6 A0A1B6L3D9 A0A151WSS5 A0A2J7Q8G9 A0A195CPX0 A0A195BXH2 A0A158NWP1 A0A195FPR1 E2BKC9 F4W3T0 A0A1B0DP54 E1ZZE2 A0A067RR30 A0A182RTJ3 A0A151JQV3 A0A026W3D5 A0A182P6I5 A0A3L8DBV7 E0VU53 A0A0J7KSS4 A0A182MHQ8 A0A1L8DV35 A0A1L8DV31 A0A182WG67 A0A1B0CIX4 K7IRU2 A0A182FW63 A0A3Q0JCB3 A0A084WHJ3 A0A2S2PE55 A0A232FHF5 A0A182USD3 A0A182TM66 A0A182Y1P0 A0A182KSH9 A0A182WZM2 A0NGI8 A0A182HQ68 W5JJU5 A0A182S991 A0A182QB47 A0A2R7VZ36 T1HMF8 A0A182IYD8 A0A182MYN8 E9ID97 A0A023F3R7 J9KB48 Q0IEB4 A0A1S4FTH6 A0A182GZS8 A0A336MC90 B0WTE5 A0A2S2R1M2 A0A1W4WIZ0 T1J5D9 A0A1W4WTU3 B7PKR8 A0A131YXS5 A0A224Y904 V5HNP7 A0A2R5LLS7 A0A1D2N3V1 A0A0K2U676 A0A1Z5KV15 A0A226F844 A0A1S3HX72 A0A1S3HVT9 A0A3R7QIS3 A0A0L8GL05 W4XT47
A0A2H1V0S1 A0A212FA78 U4UTB7 N6TDY9 A0A1Y1M8C8 D6WYK8 A0A2P8YR10 A0A1B6IAW4 A0A1B6CFM6 A0A1B6L3D9 A0A151WSS5 A0A2J7Q8G9 A0A195CPX0 A0A195BXH2 A0A158NWP1 A0A195FPR1 E2BKC9 F4W3T0 A0A1B0DP54 E1ZZE2 A0A067RR30 A0A182RTJ3 A0A151JQV3 A0A026W3D5 A0A182P6I5 A0A3L8DBV7 E0VU53 A0A0J7KSS4 A0A182MHQ8 A0A1L8DV35 A0A1L8DV31 A0A182WG67 A0A1B0CIX4 K7IRU2 A0A182FW63 A0A3Q0JCB3 A0A084WHJ3 A0A2S2PE55 A0A232FHF5 A0A182USD3 A0A182TM66 A0A182Y1P0 A0A182KSH9 A0A182WZM2 A0NGI8 A0A182HQ68 W5JJU5 A0A182S991 A0A182QB47 A0A2R7VZ36 T1HMF8 A0A182IYD8 A0A182MYN8 E9ID97 A0A023F3R7 J9KB48 Q0IEB4 A0A1S4FTH6 A0A182GZS8 A0A336MC90 B0WTE5 A0A2S2R1M2 A0A1W4WIZ0 T1J5D9 A0A1W4WTU3 B7PKR8 A0A131YXS5 A0A224Y904 V5HNP7 A0A2R5LLS7 A0A1D2N3V1 A0A0K2U676 A0A1Z5KV15 A0A226F844 A0A1S3HX72 A0A1S3HVT9 A0A3R7QIS3 A0A0L8GL05 W4XT47
Ontologies
GO
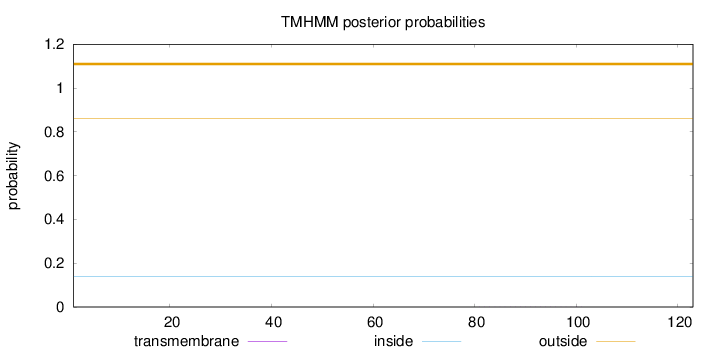
Topology
Length:
123
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00489
Exp number, first 60 AAs:
0.00089
Total prob of N-in:
0.13804
outside
1 - 123
Population Genetic Test Statistics
Pi
31.567559
Theta
29.64313
Tajima's D
0.251283
CLR
0.073367
CSRT
0.444827758612069
Interpretation
Uncertain
