Gene
KWMTBOMO04689
Pre Gene Modal
BGIBMGA009815
Annotation
PREDICTED:_mutS_protein_homolog_4-like_isoform_X1_[Bombyx_mori]
Full name
DNA mismatch repair protein
Location in the cell
Cytoplasmic Reliability : 1.737 Nuclear Reliability : 2.166
Sequence
CDS
ATGTGTTGTCTTCTTGGTGTGCTTGGCCCGACTTATACTGTTGGAGGGATTAGGGCATTGAGGGCTTCTATCTTGCAACCGTCTTGCAATAAACAATTAATTGAATCAAAACTAGATGTTCTTCAAGATTTAATTGAAAATGACAATGGTTTGATGGCTGATTTACAGGACATTTTGAAAAGGCTATCAGATATTGATAAAATATTATTCTTGTGCGTTGAGCATAATATTCAAAATGCTGACAAATTAGGAGAGGCGCAGCTCAATCAAGTGTTAATTCTAAAAACGACAATGAATATGGTACCAAAACTAGTTGAAGCTTTGAATCGTGCCGAAAGTGGTCTGTTAAGACAATTTAAAACGGATTTCGAAAATCCACATTACAATGAAATTGCCGAACGCATAAAAAATATCATACAAGACGACGCGCATTTCGAGAAGGGTGTTATGGGAAATCTCCAAAGATGTTTTGCAGTCAAGCCCGACATTAACGGACTCCTTGATGTGGCCAGAAGGACGTATTCTGAATTAATAGATGACATACAGAAAATTGTCGAACAGCTCAGCGAGACATACGACCTGCCTCTGCGGCTAAACCAAAACGTTATGAAAGGATTCCACATAGTTTTACCTATAGCTCCAAAAAACAGACGCAATTTCAGTATTGAAGATCTGCCCCCAATTTTCATACAGGTACACAACAACGGAGCCAGCGTATCGATGTCGACTGAAGAACTTGTAGTTCTCGATCAACAGGCAAAGGAATCTTTGAATGAAATACAAAGAATGAGTAACATAGTTATCGACGCATTATTGAAAGACTTAAGGCCATACATGTCGAGTCTGTACAAATTATGCGAGAACGTTGCTGAAATTGACATCCTCCTGGCATTAGCACAGGTTCGGATTGTACATATTTTAATGGAACAAAAGCATTATAACTCGTACCTCTTAACTTCTTATATGTTCCAGGCTAGCATGGTAGGGTCGTATATCAGACCGGAATTTAACAGCTATTTAGAAATAAGAAACAGCGTCCATCCGTTATTGGATTATAACAGTCACGTGATGCCAGTTCCAAATGACATAAATGCCAGCCCAGAATACAATTTCACTATAATCACCGGTCCGAATATGGGCGGCAAAAGTGTTTATATAAAACAGATTGCTATTATGCATGTTATGGCGCAACAGATTGGTTGTTTCTTGCCCGCTACTAATGCTGTACTGCGACTTAGTGACAGAATATTTTCGAGAATCGGTTTCAACGACAGCGTTGAGTTGAACGCTTCTACTTTTGTTTTCGAGATGAAAGAGATGCAGCATATTTTAAAAGGACTGACGCCATCAAGCCTGGTCATAATAGACGAACTCTGTCGCGGTACAACATACGAAGAGGGAGTCAGTATAGCTTGGGCGATCTGTGAAGAATTAATATTATCTGACGCTTTCACGTTCTTCACAACACACTTTTTATATTTGACGAAATTGCAACACCTTTACTACAACGTGGTCAATCGCCATACTACTGTTAAGGAAGAAAACGTTGATGAACTGAACGATTCTCATAAAAAACTGATTTATGAACATAAAATAGAATTAGGAGTTACGAATATAAAAAACTATGGATTGGCTTTGGCAGCAAAAACAAATTTACCAGAAGACACCGTAAAACTTGCATTTGAACTAGCAGAACTTATGACCATTGATGAAGAGACGCCAGAAACCTGCCTGCCGCAAAGAAATGACGAAAGAGAACTTTATATTTTAAATGCAGAGCTACAAAAAGAATATAGAAAAAATGGAGGTTCAGAGAAAGCTTTGGAAGCCACCATAAACAAATTCAAAGAAAATAATGCTGAACTCGTTGAAAAATTAAAAATTCAAACCAGCAACAATCCTTCTAGTTCAAGTAACGAATGTTGGTCAAGTGACCCTAAAAAAGGCTATTCAAATAATCCCAGTGGCAGTGGTAAAAAACCATTGGAATATAATAGAACACCCGAACGACTAAATACCGAGTCTTTAACAGTTAATGAAAAACCCGGTGCTACAGAGATATACCCACAAATCAAATCTTCCAATGAACTATTGACAAGCATGAAAACTATAAAGAACAAATCTCATACGTACATTGACGAAGACATACAAACGATTGGTATAGAAAACGTTAATACTAAATTAAATAAATTATCTCCAAATTGTTTAAATGAAAGTAATCATTACATTGAAGAATACCAAAATACGTTAAGCACGGAACATGAGAATTCACTCAGTGATGACGATTTATCAGAAGCACTAACGCAGGTTATAGGAGAATGTTCCAATTCATTCGATCATTCAATAGATAACGAGGATAGGGCTACCGATGAAACCCTTATGATAGAAACGATTAATGAAATTAATGAGGAATTGAACAATGCATCTAATTATAATTTTAACGATTTAATTTTAACGCCACCAATCGCTTTCAGGGACATTTCGTAG
Protein
MCCLLGVLGPTYTVGGIRALRASILQPSCNKQLIESKLDVLQDLIENDNGLMADLQDILKRLSDIDKILFLCVEHNIQNADKLGEAQLNQVLILKTTMNMVPKLVEALNRAESGLLRQFKTDFENPHYNEIAERIKNIIQDDAHFEKGVMGNLQRCFAVKPDINGLLDVARRTYSELIDDIQKIVEQLSETYDLPLRLNQNVMKGFHIVLPIAPKNRRNFSIEDLPPIFIQVHNNGASVSMSTEELVVLDQQAKESLNEIQRMSNIVIDALLKDLRPYMSSLYKLCENVAEIDILLALAQVRIVHILMEQKHYNSYLLTSYMFQASMVGSYIRPEFNSYLEIRNSVHPLLDYNSHVMPVPNDINASPEYNFTIITGPNMGGKSVYIKQIAIMHVMAQQIGCFLPATNAVLRLSDRIFSRIGFNDSVELNASTFVFEMKEMQHILKGLTPSSLVIIDELCRGTTYEEGVSIAWAICEELILSDAFTFFTTHFLYLTKLQHLYYNVVNRHTTVKEENVDELNDSHKKLIYEHKIELGVTNIKNYGLALAAKTNLPEDTVKLAFELAELMTIDEETPETCLPQRNDERELYILNAELQKEYRKNGGSEKALEATINKFKENNAELVEKLKIQTSNNPSSSSNECWSSDPKKGYSNNPSGSGKKPLEYNRTPERLNTESLTVNEKPGATEIYPQIKSSNELLTSMKTIKNKSHTYIDEDIQTIGIENVNTKLNKLSPNCLNESNHYIEEYQNTLSTEHENSLSDDDLSEALTQVIGECSNSFDHSIDNEDRATDETLMIETINEINEELNNASNYNFNDLILTPPIAFRDIS
Summary
Description
Component of the post-replicative DNA mismatch repair system (MMR).
Similarity
Belongs to the DNA mismatch repair MutS family.
Uniprot
H9JJW4
A0A2H1VXG5
A0A194PQU0
A0A194QYE7
A0A2A4IVB4
A0A3S2P2U8
+ More
A0A0L7KVA3 A0A1S3J8M3 A0A291S6W3 A0A0L8GSV3 A0A310S973 R7UWW9 X1X5V3 A0A210Q4E9 A0A087ZW46 A0A2T7PYR7 G3NYW7 C3Z1V1 A0A3Q3J7J5 A0A3B3Y865 A0A315V2X0 A0A3Q2XGU5 X1WBE0 A0A3Q1JBY1 A0A3B3V842 A0A3Q2PPU5 A0A2I4BLW9 A0A087XAI6 A0A2Y9D7G1 A0A151NJZ5 A7SM04 A0A3B5ALJ5 A0A3B4T9K8 A0A091CZD1 W5MZK1 F1P6X2 A0A1W5AVU3 A0A1U7T652 A0A2U3XQC5 A0A091Q636 A0A087WQN0 A0A3B4YRE2 F1M9U4 A0A091SRK3 A0A3Q0GHF2 M3YHF4 A0A0Q3MKY6 I3MR69 H0UVG2 A0A093JB45 A0A3P9J148 A0A2I2USD9 A0A093NKS1 A0A3Q7TGP9 A0A3Q0DCQ3 H3AEP5 F6TF84 A0A3B5QL95 Q7TNA6 E2RJA9 M3XLC8 A0A091TZC5 A0A3Q1EZY6 A0A087WQG8 U3JF60 A0A091HJ69 A0A2Y9IWB1 A0A0A0AD33 A0A2Y9G427 H0WWT9 A0A099ZCQ0 H3DJ20 F6QL61
A0A0L7KVA3 A0A1S3J8M3 A0A291S6W3 A0A0L8GSV3 A0A310S973 R7UWW9 X1X5V3 A0A210Q4E9 A0A087ZW46 A0A2T7PYR7 G3NYW7 C3Z1V1 A0A3Q3J7J5 A0A3B3Y865 A0A315V2X0 A0A3Q2XGU5 X1WBE0 A0A3Q1JBY1 A0A3B3V842 A0A3Q2PPU5 A0A2I4BLW9 A0A087XAI6 A0A2Y9D7G1 A0A151NJZ5 A7SM04 A0A3B5ALJ5 A0A3B4T9K8 A0A091CZD1 W5MZK1 F1P6X2 A0A1W5AVU3 A0A1U7T652 A0A2U3XQC5 A0A091Q636 A0A087WQN0 A0A3B4YRE2 F1M9U4 A0A091SRK3 A0A3Q0GHF2 M3YHF4 A0A0Q3MKY6 I3MR69 H0UVG2 A0A093JB45 A0A3P9J148 A0A2I2USD9 A0A093NKS1 A0A3Q7TGP9 A0A3Q0DCQ3 H3AEP5 F6TF84 A0A3B5QL95 Q7TNA6 E2RJA9 M3XLC8 A0A091TZC5 A0A3Q1EZY6 A0A087WQG8 U3JF60 A0A091HJ69 A0A2Y9IWB1 A0A0A0AD33 A0A2Y9G427 H0WWT9 A0A099ZCQ0 H3DJ20 F6QL61
Pubmed
EMBL
BABH01040635
ODYU01004714
SOQ44894.1
KQ459595
KPI95677.1
KQ461154
+ More
KPJ08601.1 NWSH01006823 PCG63224.1 RSAL01000041 RVE50926.1 JTDY01005259 KOB67187.1 MF432997 ATL75348.1 KQ420579 KOF79909.1 KQ763441 OAD55100.1 AMQN01005970 KB297235 ELU10757.1 ABLF02007369 ABLF02007370 ABLF02007371 ABLF02054782 NEDP02005059 OWF43559.1 PZQS01000001 PVD38561.1 GG666573 EEN53468.1 NHOQ01002481 PWA16448.1 BX465227 AYCK01005147 AKHW03002907 KYO36835.1 DS469703 EDO35291.1 KN123809 KFO23250.1 AHAT01003728 AAEX03004895 AAEX03004896 KK687050 KFQ15001.1 AC087184 AABR07013859 AABR07013860 KK483858 KFQ61347.1 AEYP01045765 AEYP01045766 AEYP01045767 AEYP01045768 AEYP01045769 AEYP01045770 AEYP01045771 AEYP01045772 LMAW01001570 KQK83168.1 AGTP01063383 AGTP01063384 AGTP01063385 AAKN02005014 AAKN02005015 KL217284 KFV76134.1 AANG04003279 KL224841 KFW64726.1 AFYH01155080 AFYH01155081 AFYH01155082 AFYH01155083 AFYH01155084 JSUE03001697 JSUE03001698 JSUE03001699 AY351586 AAQ20789.1 KK407711 KFQ83127.1 AGTO01011277 KL217539 KFO96338.1 KL871554 KGL92419.1 AAQR03023681 AAQR03023682 AAQR03023683 AAQR03023684 AAQR03023685 AAQR03023686 AAQR03023687 KL891373 KGL78610.1
KPJ08601.1 NWSH01006823 PCG63224.1 RSAL01000041 RVE50926.1 JTDY01005259 KOB67187.1 MF432997 ATL75348.1 KQ420579 KOF79909.1 KQ763441 OAD55100.1 AMQN01005970 KB297235 ELU10757.1 ABLF02007369 ABLF02007370 ABLF02007371 ABLF02054782 NEDP02005059 OWF43559.1 PZQS01000001 PVD38561.1 GG666573 EEN53468.1 NHOQ01002481 PWA16448.1 BX465227 AYCK01005147 AKHW03002907 KYO36835.1 DS469703 EDO35291.1 KN123809 KFO23250.1 AHAT01003728 AAEX03004895 AAEX03004896 KK687050 KFQ15001.1 AC087184 AABR07013859 AABR07013860 KK483858 KFQ61347.1 AEYP01045765 AEYP01045766 AEYP01045767 AEYP01045768 AEYP01045769 AEYP01045770 AEYP01045771 AEYP01045772 LMAW01001570 KQK83168.1 AGTP01063383 AGTP01063384 AGTP01063385 AAKN02005014 AAKN02005015 KL217284 KFV76134.1 AANG04003279 KL224841 KFW64726.1 AFYH01155080 AFYH01155081 AFYH01155082 AFYH01155083 AFYH01155084 JSUE03001697 JSUE03001698 JSUE03001699 AY351586 AAQ20789.1 KK407711 KFQ83127.1 AGTO01011277 KL217539 KFO96338.1 KL871554 KGL92419.1 AAQR03023681 AAQR03023682 AAQR03023683 AAQR03023684 AAQR03023685 AAQR03023686 AAQR03023687 KL891373 KGL78610.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000283053
UP000037510
+ More
UP000085678 UP000053454 UP000014760 UP000007819 UP000242188 UP000005203 UP000245119 UP000007635 UP000001554 UP000261600 UP000261480 UP000264820 UP000000437 UP000265040 UP000261500 UP000265000 UP000192220 UP000028760 UP000248480 UP000050525 UP000001593 UP000261400 UP000261420 UP000028990 UP000018468 UP000002254 UP000192224 UP000189704 UP000245341 UP000000589 UP000261360 UP000002494 UP000189705 UP000000715 UP000051836 UP000005215 UP000005447 UP000053875 UP000265200 UP000011712 UP000054081 UP000286640 UP000189706 UP000008672 UP000006718 UP000002852 UP000257200 UP000016665 UP000054308 UP000248482 UP000053858 UP000248481 UP000005225 UP000053641 UP000007303 UP000008225
UP000085678 UP000053454 UP000014760 UP000007819 UP000242188 UP000005203 UP000245119 UP000007635 UP000001554 UP000261600 UP000261480 UP000264820 UP000000437 UP000265040 UP000261500 UP000265000 UP000192220 UP000028760 UP000248480 UP000050525 UP000001593 UP000261400 UP000261420 UP000028990 UP000018468 UP000002254 UP000192224 UP000189704 UP000245341 UP000000589 UP000261360 UP000002494 UP000189705 UP000000715 UP000051836 UP000005215 UP000005447 UP000053875 UP000265200 UP000011712 UP000054081 UP000286640 UP000189706 UP000008672 UP000006718 UP000002852 UP000257200 UP000016665 UP000054308 UP000248482 UP000053858 UP000248481 UP000005225 UP000053641 UP000007303 UP000008225
PRIDE
Interpro
IPR000432
DNA_mismatch_repair_MutS_C
+ More
IPR036678 MutS_con_dom_sf
IPR011184 DNA_mismatch_repair_Msh2
IPR007696 DNA_mismatch_repair_MutS_core
IPR036187 DNA_mismatch_repair_MutS_sf
IPR027417 P-loop_NTPase
IPR007860 DNA_mmatch_repair_MutS_con_dom
IPR007861 DNA_mismatch_repair_MutS_clamp
IPR017261 DNA_mismatch_repair_MutS/MSH
IPR036678 MutS_con_dom_sf
IPR011184 DNA_mismatch_repair_Msh2
IPR007696 DNA_mismatch_repair_MutS_core
IPR036187 DNA_mismatch_repair_MutS_sf
IPR027417 P-loop_NTPase
IPR007860 DNA_mmatch_repair_MutS_con_dom
IPR007861 DNA_mismatch_repair_MutS_clamp
IPR017261 DNA_mismatch_repair_MutS/MSH
Gene 3D
ProteinModelPortal
H9JJW4
A0A2H1VXG5
A0A194PQU0
A0A194QYE7
A0A2A4IVB4
A0A3S2P2U8
+ More
A0A0L7KVA3 A0A1S3J8M3 A0A291S6W3 A0A0L8GSV3 A0A310S973 R7UWW9 X1X5V3 A0A210Q4E9 A0A087ZW46 A0A2T7PYR7 G3NYW7 C3Z1V1 A0A3Q3J7J5 A0A3B3Y865 A0A315V2X0 A0A3Q2XGU5 X1WBE0 A0A3Q1JBY1 A0A3B3V842 A0A3Q2PPU5 A0A2I4BLW9 A0A087XAI6 A0A2Y9D7G1 A0A151NJZ5 A7SM04 A0A3B5ALJ5 A0A3B4T9K8 A0A091CZD1 W5MZK1 F1P6X2 A0A1W5AVU3 A0A1U7T652 A0A2U3XQC5 A0A091Q636 A0A087WQN0 A0A3B4YRE2 F1M9U4 A0A091SRK3 A0A3Q0GHF2 M3YHF4 A0A0Q3MKY6 I3MR69 H0UVG2 A0A093JB45 A0A3P9J148 A0A2I2USD9 A0A093NKS1 A0A3Q7TGP9 A0A3Q0DCQ3 H3AEP5 F6TF84 A0A3B5QL95 Q7TNA6 E2RJA9 M3XLC8 A0A091TZC5 A0A3Q1EZY6 A0A087WQG8 U3JF60 A0A091HJ69 A0A2Y9IWB1 A0A0A0AD33 A0A2Y9G427 H0WWT9 A0A099ZCQ0 H3DJ20 F6QL61
A0A0L7KVA3 A0A1S3J8M3 A0A291S6W3 A0A0L8GSV3 A0A310S973 R7UWW9 X1X5V3 A0A210Q4E9 A0A087ZW46 A0A2T7PYR7 G3NYW7 C3Z1V1 A0A3Q3J7J5 A0A3B3Y865 A0A315V2X0 A0A3Q2XGU5 X1WBE0 A0A3Q1JBY1 A0A3B3V842 A0A3Q2PPU5 A0A2I4BLW9 A0A087XAI6 A0A2Y9D7G1 A0A151NJZ5 A7SM04 A0A3B5ALJ5 A0A3B4T9K8 A0A091CZD1 W5MZK1 F1P6X2 A0A1W5AVU3 A0A1U7T652 A0A2U3XQC5 A0A091Q636 A0A087WQN0 A0A3B4YRE2 F1M9U4 A0A091SRK3 A0A3Q0GHF2 M3YHF4 A0A0Q3MKY6 I3MR69 H0UVG2 A0A093JB45 A0A3P9J148 A0A2I2USD9 A0A093NKS1 A0A3Q7TGP9 A0A3Q0DCQ3 H3AEP5 F6TF84 A0A3B5QL95 Q7TNA6 E2RJA9 M3XLC8 A0A091TZC5 A0A3Q1EZY6 A0A087WQG8 U3JF60 A0A091HJ69 A0A2Y9IWB1 A0A0A0AD33 A0A2Y9G427 H0WWT9 A0A099ZCQ0 H3DJ20 F6QL61
PDB
1FW6
E-value=1.81595e-34,
Score=368
Ontologies
GO
Topology
Subcellular location
Nucleus
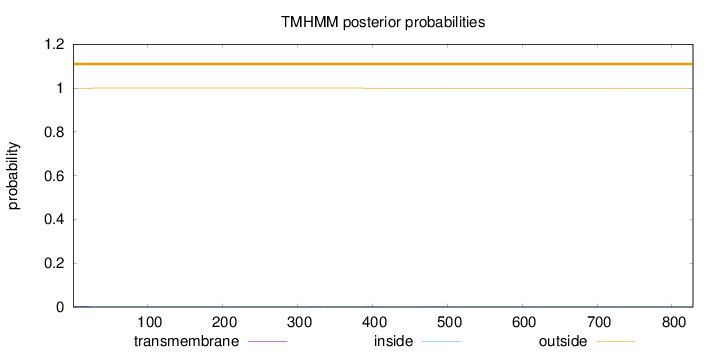
Length:
828
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10579
Exp number, first 60 AAs:
0.07268
Total prob of N-in:
0.00370
outside
1 - 828
Population Genetic Test Statistics
Pi
179.959134
Theta
169.287557
Tajima's D
0.305681
CLR
0.133867
CSRT
0.459027048647568
Interpretation
Uncertain
