Pre Gene Modal
BGIBMGA009814
Annotation
PREDICTED:_phosphoserine_phosphatase_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.693
Sequence
CDS
ATGGCTCTTTACAGTTTGAAAAGCAACCTGCAGTACGCGACGTTGAAGACGCTCTCACTGGTGTCGATCAGCGTGATGTCGCCCCAACAGACTGTCCAGGAGTTGTTCAGGACAGCGGACTGCGTTTGCTTCGATGTAGACTCGACCGTCATACAAGATGAAGGCATCGATGAACTGGCCAAGTTCTGCGGGAAAGGAGACGAGGTTAAAAGACTGACGGCGGAAGCAATGGGCGGCAATATGACATTCCAGGAAGCCTTGAAGAAGAGACTGGACATCATCAGGCCTAACGTCGGTCAGATCAGAGAATTCATTGAGAAATTTCCAGTTAGACTAACTCCTGGTATTACAGAGTTAGTGAAAGAGTTACACGAAAGAGGAGTCATCGTATATCTAGTTTCGGGCGGATTCAGGAGTCTAATAGAACCGGTCGCGGAGAGACTAAACATCCCAACTATCAACATCTTCGCTAATCGACTCAAGTTTTATTTCAATGGTGAATACGCTGGCTTCGATGAAAACGAGCCAACATCGAGATCTGGTGGCAAAGGTTTAGTGGTGAGACGACTGAAGGAATTGCACGGGTACCAGCGCCTGGTGATCGTCGGGGACGGGGCTACCGATGCCGAGGCTAGCCCACCTGCTGACGGCTTCATTGGCTTCGGGGGTAACGTGGTGCGTGACGAAGTCAAGAAGAAGGCAGCCTGGTACGTCACGGACTTCCAAGAGCTGATAGATTCATTCACCATACAGACCAAGTCTTAG
Protein
MALYSLKSNLQYATLKTLSLVSISVMSPQQTVQELFRTADCVCFDVDSTVIQDEGIDELAKFCGKGDEVKRLTAEAMGGNMTFQEALKKRLDIIRPNVGQIREFIEKFPVRLTPGITELVKELHERGVIVYLVSGGFRSLIEPVAERLNIPTINIFANRLKFYFNGEYAGFDENEPTSRSGGKGLVVRRLKELHGYQRLVIVGDGATDAEASPPADGFIGFGGNVVRDEVKKKAAWYVTDFQELIDSFTIQTKS
Summary
Uniprot
A0A3S2NLS5
A0A3G1T178
A0A2H1VVQ1
A0A194PQF1
A0JCK1
C6K7H8
+ More
A0A0L7L7A7 A0A1B6I462 A0A1B6FIB9 A0A1B6MUJ3 A0A2J7R7G6 A0A1B6KVH6 A0A067R3T4 A0A1B6FKH0 A0A1B6I3M1 L7WSE2 A0A1D2NFM6 D2A0T2 A0A0C9RBC7 A0A0T6AWF8 A0A226ER59 M7BYX9 A0A093IIE3 A0A0S7IBP8 A0A146WFF6 A0A091Q4U7 A0A3Q2CM41 A0A2D4J985 A0A091JJG3 A0A093QR11 A0A0S7I7X0 A0A2H6N5W8 A0A2D4K9Q5 A0A2H6N5Y7 A0A3Q2QAU1 A0A091HTK0 A0A091UKL6 A0A3Q2NY01 A0A3P9QEY3 A0A093HT74 A0A093R7F7 A0A3B4B9C6 A0A091QNA0 A0A3M0KC53 A0A093GRQ6 A0A0B8RPP4 A0A093BJQ2 A0A3B3YN85 A0A091ULQ3 R9QZD8 A0A091F237 H0Z3V0 A0A091W675 A0A087VKQ0 A0A0A0AK12 A0A093NC65 A0A087QK46 E1C9F4 R0LQ88 A0A218V2J9 U3JWC0 A0A099YU43 K7G1X4 A0A0Q3PDM3 A0A093LL54 A0A3L8RYE0 G1MYR6 A0A3Q3B969 A0A091HUX7 A0A091UE14 A0A1V4K2U0 A0A2P4TGV9 A0A3B3V387 A0A1U7SCR8 A0A093JT82 A0A2I0M1Q0 A0A0K8SQR0 A0A1U7SMS1 A0A3B3XAM1 A0A315VBY6 A0A147A362 A0A146M672 A0A091GK59 Q0IHZ7 A0A2H8TM38 A0A2S2NJW6 A0A0A9YW53 Q6DFU1 A0A0K8SR14 A0A2J7R7G0 A0A091PIA1 A0A1L8HG72 A0A3B5M0Y2 A0A1W7RET6 T1DJQ2 A0A087YC19 A0A3B3V3M2 A0A3P8VLI6 C4WU94
A0A0L7L7A7 A0A1B6I462 A0A1B6FIB9 A0A1B6MUJ3 A0A2J7R7G6 A0A1B6KVH6 A0A067R3T4 A0A1B6FKH0 A0A1B6I3M1 L7WSE2 A0A1D2NFM6 D2A0T2 A0A0C9RBC7 A0A0T6AWF8 A0A226ER59 M7BYX9 A0A093IIE3 A0A0S7IBP8 A0A146WFF6 A0A091Q4U7 A0A3Q2CM41 A0A2D4J985 A0A091JJG3 A0A093QR11 A0A0S7I7X0 A0A2H6N5W8 A0A2D4K9Q5 A0A2H6N5Y7 A0A3Q2QAU1 A0A091HTK0 A0A091UKL6 A0A3Q2NY01 A0A3P9QEY3 A0A093HT74 A0A093R7F7 A0A3B4B9C6 A0A091QNA0 A0A3M0KC53 A0A093GRQ6 A0A0B8RPP4 A0A093BJQ2 A0A3B3YN85 A0A091ULQ3 R9QZD8 A0A091F237 H0Z3V0 A0A091W675 A0A087VKQ0 A0A0A0AK12 A0A093NC65 A0A087QK46 E1C9F4 R0LQ88 A0A218V2J9 U3JWC0 A0A099YU43 K7G1X4 A0A0Q3PDM3 A0A093LL54 A0A3L8RYE0 G1MYR6 A0A3Q3B969 A0A091HUX7 A0A091UE14 A0A1V4K2U0 A0A2P4TGV9 A0A3B3V387 A0A1U7SCR8 A0A093JT82 A0A2I0M1Q0 A0A0K8SQR0 A0A1U7SMS1 A0A3B3XAM1 A0A315VBY6 A0A147A362 A0A146M672 A0A091GK59 Q0IHZ7 A0A2H8TM38 A0A2S2NJW6 A0A0A9YW53 Q6DFU1 A0A0K8SR14 A0A2J7R7G0 A0A091PIA1 A0A1L8HG72 A0A3B5M0Y2 A0A1W7RET6 T1DJQ2 A0A087YC19 A0A3B3V3M2 A0A3P8VLI6 C4WU94
Pubmed
EMBL
RSAL01000041
RVE50925.1
MG846871
AXY94723.1
ODYU01004714
SOQ44893.1
+ More
KQ459595 KPI95676.1 AB282639 BAF36819.1 GQ166954 ACS34989.1 JTDY01002573 KOB71166.1 GECU01026026 JAS81680.1 GECZ01030351 GECZ01020066 JAS39418.1 JAS49703.1 GEBQ01000396 JAT39581.1 NEVH01006732 PNF36771.1 GEBQ01024586 GEBQ01016786 JAT15391.1 JAT23191.1 KK853008 KDR12562.1 GECZ01019276 JAS50493.1 GECU01026168 JAS81538.1 KC118985 AGC84407.1 LJIJ01000056 ODN04078.1 KQ971338 EFA01638.1 GBYB01005625 JAG75392.1 LJIG01022645 KRT79466.1 LNIX01000002 OXA59524.1 KB499736 EMP41080.1 KK562859 KFW02475.1 GBYX01432052 JAO49261.1 GCES01057550 JAR28773.1 KK687582 KFQ15222.1 IACK01158566 LAA93047.1 KK502172 KFP21154.1 KL672574 KFW86407.1 GBYX01432053 JAO49260.1 IACI01052874 LAA25276.1 IACL01036719 LAB05438.1 IACI01052873 LAA25275.1 KL217786 KFO98479.1 KL409726 KFQ91479.1 KK396852 KFV57728.1 KL442409 KFW94535.1 KK800095 KFQ28101.1 QRBI01000111 RMC10786.1 KL216427 KFV69479.1 GBSH01001977 JAG67049.1 KN126073 KFU86350.1 KK457129 KFQ77013.1 JX125596 AGG09860.1 KK719555 KFO64213.1 ABQF01026247 KK734748 KFR10615.1 KL496961 KFO13192.1 KL871755 KGL93385.1 KL224666 KFW62528.1 KL225707 KFM01600.1 AADN05000057 KB742489 EOB07869.1 MUZQ01000070 OWK59950.1 AGTO01000878 KL886357 KGL73794.1 AGCU01019389 LMAW01003051 KQK74372.1 KK606820 KFW09606.1 QUSF01000121 RLV90575.1 KL525625 KFO91053.1 KK430063 KFQ88761.1 LSYS01005108 OPJ78691.1 PPHD01000366 POI35564.1 KL206367 KFV81799.1 AKCR02000048 PKK23606.1 GBRD01017779 GBRD01010263 GBRD01010259 GDHC01009101 JAG55561.1 JAQ09528.1 NHOQ01001971 PWA20326.1 GCES01013333 JAR72990.1 GDHC01003690 JAQ14939.1 KL447524 KFO74512.1 AAMC01050234 BC122888 BC158947 AAI22889.1 AAI58948.1 GFXV01003206 MBW15011.1 GGMR01004855 MBY17474.1 GBHO01006372 GBRD01010261 GDHC01020433 GDHC01010992 JAG37232.1 JAG55563.1 JAP98195.1 JAQ07637.1 BC076642 AAH76642.1 GBRD01010265 GBRD01010264 GBRD01010260 JAG55559.1 PNF36774.1 KK659731 KFQ07385.1 CM004468 OCT95086.1 GDAY02001770 JAV49658.1 GAAZ01001793 JAA96150.1 AYCK01017765 AK340971 BAH71464.1
KQ459595 KPI95676.1 AB282639 BAF36819.1 GQ166954 ACS34989.1 JTDY01002573 KOB71166.1 GECU01026026 JAS81680.1 GECZ01030351 GECZ01020066 JAS39418.1 JAS49703.1 GEBQ01000396 JAT39581.1 NEVH01006732 PNF36771.1 GEBQ01024586 GEBQ01016786 JAT15391.1 JAT23191.1 KK853008 KDR12562.1 GECZ01019276 JAS50493.1 GECU01026168 JAS81538.1 KC118985 AGC84407.1 LJIJ01000056 ODN04078.1 KQ971338 EFA01638.1 GBYB01005625 JAG75392.1 LJIG01022645 KRT79466.1 LNIX01000002 OXA59524.1 KB499736 EMP41080.1 KK562859 KFW02475.1 GBYX01432052 JAO49261.1 GCES01057550 JAR28773.1 KK687582 KFQ15222.1 IACK01158566 LAA93047.1 KK502172 KFP21154.1 KL672574 KFW86407.1 GBYX01432053 JAO49260.1 IACI01052874 LAA25276.1 IACL01036719 LAB05438.1 IACI01052873 LAA25275.1 KL217786 KFO98479.1 KL409726 KFQ91479.1 KK396852 KFV57728.1 KL442409 KFW94535.1 KK800095 KFQ28101.1 QRBI01000111 RMC10786.1 KL216427 KFV69479.1 GBSH01001977 JAG67049.1 KN126073 KFU86350.1 KK457129 KFQ77013.1 JX125596 AGG09860.1 KK719555 KFO64213.1 ABQF01026247 KK734748 KFR10615.1 KL496961 KFO13192.1 KL871755 KGL93385.1 KL224666 KFW62528.1 KL225707 KFM01600.1 AADN05000057 KB742489 EOB07869.1 MUZQ01000070 OWK59950.1 AGTO01000878 KL886357 KGL73794.1 AGCU01019389 LMAW01003051 KQK74372.1 KK606820 KFW09606.1 QUSF01000121 RLV90575.1 KL525625 KFO91053.1 KK430063 KFQ88761.1 LSYS01005108 OPJ78691.1 PPHD01000366 POI35564.1 KL206367 KFV81799.1 AKCR02000048 PKK23606.1 GBRD01017779 GBRD01010263 GBRD01010259 GDHC01009101 JAG55561.1 JAQ09528.1 NHOQ01001971 PWA20326.1 GCES01013333 JAR72990.1 GDHC01003690 JAQ14939.1 KL447524 KFO74512.1 AAMC01050234 BC122888 BC158947 AAI22889.1 AAI58948.1 GFXV01003206 MBW15011.1 GGMR01004855 MBY17474.1 GBHO01006372 GBRD01010261 GDHC01020433 GDHC01010992 JAG37232.1 JAG55563.1 JAP98195.1 JAQ07637.1 BC076642 AAH76642.1 GBRD01010265 GBRD01010264 GBRD01010260 JAG55559.1 PNF36774.1 KK659731 KFQ07385.1 CM004468 OCT95086.1 GDAY02001770 JAV49658.1 GAAZ01001793 JAA96150.1 AYCK01017765 AK340971 BAH71464.1
Proteomes
UP000283053
UP000053268
UP000037510
UP000235965
UP000027135
UP000094527
+ More
UP000007266 UP000198287 UP000031443 UP000265020 UP000053119 UP000053258 UP000265000 UP000054308 UP000053283 UP000242638 UP000261520 UP000269221 UP000053875 UP000261480 UP000052976 UP000007754 UP000053605 UP000053858 UP000054081 UP000053286 UP000000539 UP000197619 UP000016665 UP000053641 UP000007267 UP000051836 UP000276834 UP000001645 UP000264800 UP000190648 UP000261500 UP000189705 UP000053584 UP000053872 UP000053760 UP000008143 UP000186698 UP000261380 UP000028760 UP000265120
UP000007266 UP000198287 UP000031443 UP000265020 UP000053119 UP000053258 UP000265000 UP000054308 UP000053283 UP000242638 UP000261520 UP000269221 UP000053875 UP000261480 UP000052976 UP000007754 UP000053605 UP000053858 UP000054081 UP000053286 UP000000539 UP000197619 UP000016665 UP000053641 UP000007267 UP000051836 UP000276834 UP000001645 UP000264800 UP000190648 UP000261500 UP000189705 UP000053584 UP000053872 UP000053760 UP000008143 UP000186698 UP000261380 UP000028760 UP000265120
SUPFAM
SSF56784
SSF56784
Gene 3D
ProteinModelPortal
A0A3S2NLS5
A0A3G1T178
A0A2H1VVQ1
A0A194PQF1
A0JCK1
C6K7H8
+ More
A0A0L7L7A7 A0A1B6I462 A0A1B6FIB9 A0A1B6MUJ3 A0A2J7R7G6 A0A1B6KVH6 A0A067R3T4 A0A1B6FKH0 A0A1B6I3M1 L7WSE2 A0A1D2NFM6 D2A0T2 A0A0C9RBC7 A0A0T6AWF8 A0A226ER59 M7BYX9 A0A093IIE3 A0A0S7IBP8 A0A146WFF6 A0A091Q4U7 A0A3Q2CM41 A0A2D4J985 A0A091JJG3 A0A093QR11 A0A0S7I7X0 A0A2H6N5W8 A0A2D4K9Q5 A0A2H6N5Y7 A0A3Q2QAU1 A0A091HTK0 A0A091UKL6 A0A3Q2NY01 A0A3P9QEY3 A0A093HT74 A0A093R7F7 A0A3B4B9C6 A0A091QNA0 A0A3M0KC53 A0A093GRQ6 A0A0B8RPP4 A0A093BJQ2 A0A3B3YN85 A0A091ULQ3 R9QZD8 A0A091F237 H0Z3V0 A0A091W675 A0A087VKQ0 A0A0A0AK12 A0A093NC65 A0A087QK46 E1C9F4 R0LQ88 A0A218V2J9 U3JWC0 A0A099YU43 K7G1X4 A0A0Q3PDM3 A0A093LL54 A0A3L8RYE0 G1MYR6 A0A3Q3B969 A0A091HUX7 A0A091UE14 A0A1V4K2U0 A0A2P4TGV9 A0A3B3V387 A0A1U7SCR8 A0A093JT82 A0A2I0M1Q0 A0A0K8SQR0 A0A1U7SMS1 A0A3B3XAM1 A0A315VBY6 A0A147A362 A0A146M672 A0A091GK59 Q0IHZ7 A0A2H8TM38 A0A2S2NJW6 A0A0A9YW53 Q6DFU1 A0A0K8SR14 A0A2J7R7G0 A0A091PIA1 A0A1L8HG72 A0A3B5M0Y2 A0A1W7RET6 T1DJQ2 A0A087YC19 A0A3B3V3M2 A0A3P8VLI6 C4WU94
A0A0L7L7A7 A0A1B6I462 A0A1B6FIB9 A0A1B6MUJ3 A0A2J7R7G6 A0A1B6KVH6 A0A067R3T4 A0A1B6FKH0 A0A1B6I3M1 L7WSE2 A0A1D2NFM6 D2A0T2 A0A0C9RBC7 A0A0T6AWF8 A0A226ER59 M7BYX9 A0A093IIE3 A0A0S7IBP8 A0A146WFF6 A0A091Q4U7 A0A3Q2CM41 A0A2D4J985 A0A091JJG3 A0A093QR11 A0A0S7I7X0 A0A2H6N5W8 A0A2D4K9Q5 A0A2H6N5Y7 A0A3Q2QAU1 A0A091HTK0 A0A091UKL6 A0A3Q2NY01 A0A3P9QEY3 A0A093HT74 A0A093R7F7 A0A3B4B9C6 A0A091QNA0 A0A3M0KC53 A0A093GRQ6 A0A0B8RPP4 A0A093BJQ2 A0A3B3YN85 A0A091ULQ3 R9QZD8 A0A091F237 H0Z3V0 A0A091W675 A0A087VKQ0 A0A0A0AK12 A0A093NC65 A0A087QK46 E1C9F4 R0LQ88 A0A218V2J9 U3JWC0 A0A099YU43 K7G1X4 A0A0Q3PDM3 A0A093LL54 A0A3L8RYE0 G1MYR6 A0A3Q3B969 A0A091HUX7 A0A091UE14 A0A1V4K2U0 A0A2P4TGV9 A0A3B3V387 A0A1U7SCR8 A0A093JT82 A0A2I0M1Q0 A0A0K8SQR0 A0A1U7SMS1 A0A3B3XAM1 A0A315VBY6 A0A147A362 A0A146M672 A0A091GK59 Q0IHZ7 A0A2H8TM38 A0A2S2NJW6 A0A0A9YW53 Q6DFU1 A0A0K8SR14 A0A2J7R7G0 A0A091PIA1 A0A1L8HG72 A0A3B5M0Y2 A0A1W7RET6 T1DJQ2 A0A087YC19 A0A3B3V3M2 A0A3P8VLI6 C4WU94
PDB
1NNL
E-value=1.46121e-74,
Score=709
Ontologies
PATHWAY
GO
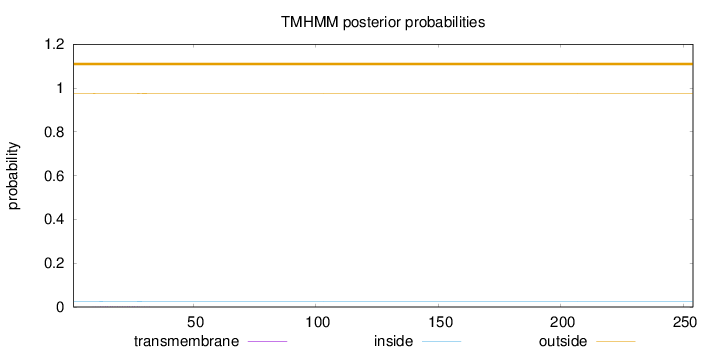
Topology
Length:
254
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02987
Exp number, first 60 AAs:
0.02786
Total prob of N-in:
0.02533
outside
1 - 254
Population Genetic Test Statistics
Pi
271.837644
Theta
187.740566
Tajima's D
1.543419
CLR
0.384469
CSRT
0.800209989500525
Interpretation
Uncertain
