Gene
KWMTBOMO04684
Pre Gene Modal
BGIBMGA005524
Annotation
PREDICTED:_UNC93-like_protein_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.952
Sequence
CDS
ATGCCGGAAGAAACAATAACTTTTAGTTCAAATGAGAAATGGCGTATAAACCGGAACGTATTCGTTCTTGGTTTCGCTTTCATGATCCACTTCACGGCCTTCCACGGGACCGCCAACCTCCAGAGCTCCGTGAACAGCGACGACGGAGCCGGCACCTTCACCCTGGCCGCCATTTACTTCTCCCTGATCCTCTCGAATATATTATTACCCGCTGTTGTCATTAAGTGGCTTGGTTGCAAATGGACAGTGTCAATGTCCTTCATCACGTATATGCCGTTCATAGCAGCGCAGTTTTATCCGAAGTTGTATACACTGATACCAGCCGGCATGATGGTCGGCTTTGGTGGCGGACCACTCTGGTGCGCTAAATGCACTTATCTTTCTGTGATATCCGAACCGTATTCTAAATTATCAGGAATTCCAGCTGAAGTTTTGGTGGTGAGGTTCTTCGGGCTGTTCTTCATGTTCTACCAGATGGCGCAAATATGGGGAAATCTTATATCTTCTGCGATTTTATCGTCATCTCTAGGAGAGGCGCCGTTTTTGAACGAAACTATTCCGTCTACTGCGATGACGCTGATCACTAACGCCTCCGAAATACCGCCTCTTGACTTTAGAGCCACATGCGGAGCTAACTTCTGCAGCAGCAGTGCTTCACACGGTAACAGTAACCTCAAGCCACCTTCCACTGCGAAGATTAACTTGATAGCCGGCGTGTACCTCGCGTGCATGGCCATTGCTGTTCTTCTGGTTGCTGTAGGGGTCGACGCACTAACCAGGTACACATCAGGTCGTACTCCCGCAAGTCGCGGTAAGTCCGGTCTCCAACTCCTGGCGGTAACGCTGCGTCAGCTCAGGCACAAGTACCAGCTTCTGTTGCTGCCCGTCATAGGCTACATCGGCGCCGAACAAGCTTTCATGGCTGCGGACTTTACACAGGCTTACGTGGGCTGTGCCTACGGTATTAGTAACATTGGATTCGTGATGATCTGCTTCGGAGCTTGGAATGCAATAGCCGCGCCCTTAGCCGGAGCACTCGTCAAGACGACAGGCCGGTATCCGGTCATGGTCACTGCTCTGGTTTTAAATCTGTGTACCTTAACCGTACTGCTGCTCTGGAGACCGAACCCTGATCAGTGGCTGGTGTTCTACGGCCTCGCTGCCGTGTGGGGAGCCGCTGATGCTGTTTGGCTCGTGCAAGTTAACGCTCTATCCGGCATCCTATTTCCCGGTCACGAGGAAGCTGCCTATTCTAACTTCAGATTGTGGGAAGCGACCGGCAGTGTTCTGGCTTACGGCAGCGCTCCGTACCTTTGTGTTAGAACCAGATTGCAAATATTGCTCGGTCTATTAATAGTCGGCTTCACTGGCTACACGATCATTGAGCTAATGGAGTATAGAGTGAAGAGGGCGCACCACCCCGAGAGGAATTTCGAACTGGTCGATAAAAAAGAAGATCTGGGACCATTTTAA
Protein
MPEETITFSSNEKWRINRNVFVLGFAFMIHFTAFHGTANLQSSVNSDDGAGTFTLAAIYFSLILSNILLPAVVIKWLGCKWTVSMSFITYMPFIAAQFYPKLYTLIPAGMMVGFGGGPLWCAKCTYLSVISEPYSKLSGIPAEVLVVRFFGLFFMFYQMAQIWGNLISSAILSSSLGEAPFLNETIPSTAMTLITNASEIPPLDFRATCGANFCSSSASHGNSNLKPPSTAKINLIAGVYLACMAIAVLLVAVGVDALTRYTSGRTPASRGKSGLQLLAVTLRQLRHKYQLLLLPVIGYIGAEQAFMAADFTQAYVGCAYGISNIGFVMICFGAWNAIAAPLAGALVKTTGRYPVMVTALVLNLCTLTVLLLWRPNPDQWLVFYGLAAVWGAADAVWLVQVNALSGILFPGHEEAAYSNFRLWEATGSVLAYGSAPYLCVRTRLQILLGLLIVGFTGYTIIELMEYRVKRAHHPERNFELVDKKEDLGPF
Summary
Uniprot
H9J7N2
A0A2A4IZ62
A0A2H1V795
A0A194PX02
A0A3S2M441
A0A212FBL2
+ More
A0A194QTF7 A0A1L8D8Y9 A0A026VWA6 A0A195DKK0 A0A195F7N1 A0A0C9QYB7 J3JY45 A0A0L7LV09 A0A1Y1NE52 A0A1Y1N7J2 N6TVP6 A0A195B2U2 A0A158NNG4 A0A195D7D3 A0A154PTD3 A0A2A3EKV3 E2C1A8 A0A151WWE6 A0A088A8U6 K7J0X6 A0A0M8ZVZ2 A0A0L7R3U9 A0A232ELQ6 F4X5B7 B0W824 A0A1Q3FMX6 A0A182GYB1 Q17DP2 W5JS61 A0A182FTU0 U4UDQ6 A0A182QVN0 A0A067R4X6 A0A182J1T0 A0A182MYP9 A0A182Y1N0 A0A182SA02 A0A182P6J5 A0A182VCI3 A0A182KSH7 A0A182WG56 A0A182HQ79 A0A182WZN3 B3MFZ2 A0A2A4IZE4 A0A2H1VHH4 A0A0J9R7Y3 A0A182MMG8 A0A0J9R9A0 A0A1W4UK09 B4P2S7 B3N8Q2 A0A2J7RRG4 Q7K3V9 A0A084VI93 B7YZU1 A0A182JUQ7 A0A182RTK5 B4MQ38 A0A0K8VBR6 B4J5W3 B4HRW5 A0A212EPZ6 W8B145 A0A034VTW1 A0A182TP89 A0A0A1X942 A0A3B0IZM8 A0A2P8Z727 B4KPE0 B4LLA2 A0A0L0CQF6 A0A0R3NRL5 A0A0R3NL82 Q7Q0A1 B4GDD9 Q292J3 A0A194QSV0 A0A0M4EBJ8 A0A182G9R9 A0A1A9W8X4 A0A182GWR4 A0A182FIU1 A0A1I8NQL3 W5JS93 A0A084VQ76 A0A1A9WHR3 A0A2A3EB67 A0A1I8PPG6 A0A088A7F7 A0A067RMS8 B4NPS0 A0A182KH76 A0A3L8DGP3
A0A194QTF7 A0A1L8D8Y9 A0A026VWA6 A0A195DKK0 A0A195F7N1 A0A0C9QYB7 J3JY45 A0A0L7LV09 A0A1Y1NE52 A0A1Y1N7J2 N6TVP6 A0A195B2U2 A0A158NNG4 A0A195D7D3 A0A154PTD3 A0A2A3EKV3 E2C1A8 A0A151WWE6 A0A088A8U6 K7J0X6 A0A0M8ZVZ2 A0A0L7R3U9 A0A232ELQ6 F4X5B7 B0W824 A0A1Q3FMX6 A0A182GYB1 Q17DP2 W5JS61 A0A182FTU0 U4UDQ6 A0A182QVN0 A0A067R4X6 A0A182J1T0 A0A182MYP9 A0A182Y1N0 A0A182SA02 A0A182P6J5 A0A182VCI3 A0A182KSH7 A0A182WG56 A0A182HQ79 A0A182WZN3 B3MFZ2 A0A2A4IZE4 A0A2H1VHH4 A0A0J9R7Y3 A0A182MMG8 A0A0J9R9A0 A0A1W4UK09 B4P2S7 B3N8Q2 A0A2J7RRG4 Q7K3V9 A0A084VI93 B7YZU1 A0A182JUQ7 A0A182RTK5 B4MQ38 A0A0K8VBR6 B4J5W3 B4HRW5 A0A212EPZ6 W8B145 A0A034VTW1 A0A182TP89 A0A0A1X942 A0A3B0IZM8 A0A2P8Z727 B4KPE0 B4LLA2 A0A0L0CQF6 A0A0R3NRL5 A0A0R3NL82 Q7Q0A1 B4GDD9 Q292J3 A0A194QSV0 A0A0M4EBJ8 A0A182G9R9 A0A1A9W8X4 A0A182GWR4 A0A182FIU1 A0A1I8NQL3 W5JS93 A0A084VQ76 A0A1A9WHR3 A0A2A3EB67 A0A1I8PPG6 A0A088A7F7 A0A067RMS8 B4NPS0 A0A182KH76 A0A3L8DGP3
Pubmed
19121390
26354079
22118469
24508170
22516182
26227816
+ More
28004739 23537049 21347285 20798317 20075255 28648823 21719571 26483478 17510324 20920257 23761445 24845553 25244985 20966253 17994087 18057021 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 24495485 25348373 25830018 29403074 26108605 15632085 12364791 30249741
28004739 23537049 21347285 20798317 20075255 28648823 21719571 26483478 17510324 20920257 23761445 24845553 25244985 20966253 17994087 18057021 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 24495485 25348373 25830018 29403074 26108605 15632085 12364791 30249741
EMBL
BABH01014093
NWSH01004487
PCG65051.1
ODYU01001025
SOQ36666.1
KQ459595
+ More
KPI95670.1 RSAL01000041 RVE50921.1 AGBW02009298 OWR51126.1 KQ461154 KPJ08594.1 GFDF01011145 JAV02939.1 KK107695 EZA48042.1 KQ980765 KYN13376.1 KQ981744 KYN36192.1 GBYB01000688 GBYB01000689 GBYB01000690 JAG70455.1 JAG70456.1 JAG70457.1 BT128169 AEE63130.1 JTDY01000027 KOB79338.1 GEZM01010690 JAV93907.1 GEZM01010691 JAV93904.1 APGK01058995 APGK01058996 APGK01058997 APGK01058998 KB741292 ENN70372.1 KQ976641 KYM78798.1 ADTU01021539 KQ976749 KYN08786.1 KQ435105 KZC14684.1 KZ288230 PBC31651.1 GL451914 EFN78270.1 KQ982691 KYQ52224.1 KQ435855 KOX70659.1 KQ414661 KOC65552.1 NNAY01003515 OXU19293.1 GL888705 EGI58365.1 DS231856 EDS38486.1 GFDL01006106 JAV28939.1 JXUM01002850 KQ560146 KXJ84175.1 CH477292 EAT44533.1 ADMH02000327 ETN66941.1 KB632235 ERL90433.1 AXCN02001033 KK852890 KDR14271.1 APCN01003297 CH902619 EDV35674.1 KPU75780.1 NWSH01005059 PCG64473.1 ODYU01002414 SOQ39862.1 CM002911 KMY92252.1 KMY92253.1 AXCM01002861 KMY92254.1 CM000157 EDW89338.1 CH954177 EDV59529.2 NEVH01000606 PNF43431.1 AE013599 AY058326 AAF59099.4 AAL13555.1 ATLV01013334 KE524853 KFB37687.1 ACL83072.1 CH963849 EDW74227.1 GDHF01015996 JAI36318.1 CH916367 EDW00806.1 CH480816 EDW46928.1 AGBW02013359 OWR43531.1 GAMC01019594 JAB86961.1 GAKP01013697 JAC45255.1 GBXI01016298 GBXI01007032 GBXI01004348 JAC97993.1 JAD07260.1 JAD09944.1 OUUW01000001 SPP73705.1 PYGN01000167 PSN52296.1 CH933808 EDW10136.1 CH940648 EDW60839.1 JRES01000171 KNC33669.1 CM000071 KRT01637.1 KRT01636.1 AAAB01008986 EAA00363.3 CH479181 EDW31615.1 EAL24869.3 KPJ08593.1 CP012524 ALC40915.1 JXUM01049812 KQ561622 KXJ77978.1 JXUM01003156 KQ560154 KXJ84146.1 ADMH02000612 ETN65624.1 ATLV01015142 KE525003 KFB40120.1 KZ288296 PBC28930.1 KK852575 KDR21025.1 CH964291 EDW86510.1 QOIP01000009 RLU19038.1
KPI95670.1 RSAL01000041 RVE50921.1 AGBW02009298 OWR51126.1 KQ461154 KPJ08594.1 GFDF01011145 JAV02939.1 KK107695 EZA48042.1 KQ980765 KYN13376.1 KQ981744 KYN36192.1 GBYB01000688 GBYB01000689 GBYB01000690 JAG70455.1 JAG70456.1 JAG70457.1 BT128169 AEE63130.1 JTDY01000027 KOB79338.1 GEZM01010690 JAV93907.1 GEZM01010691 JAV93904.1 APGK01058995 APGK01058996 APGK01058997 APGK01058998 KB741292 ENN70372.1 KQ976641 KYM78798.1 ADTU01021539 KQ976749 KYN08786.1 KQ435105 KZC14684.1 KZ288230 PBC31651.1 GL451914 EFN78270.1 KQ982691 KYQ52224.1 KQ435855 KOX70659.1 KQ414661 KOC65552.1 NNAY01003515 OXU19293.1 GL888705 EGI58365.1 DS231856 EDS38486.1 GFDL01006106 JAV28939.1 JXUM01002850 KQ560146 KXJ84175.1 CH477292 EAT44533.1 ADMH02000327 ETN66941.1 KB632235 ERL90433.1 AXCN02001033 KK852890 KDR14271.1 APCN01003297 CH902619 EDV35674.1 KPU75780.1 NWSH01005059 PCG64473.1 ODYU01002414 SOQ39862.1 CM002911 KMY92252.1 KMY92253.1 AXCM01002861 KMY92254.1 CM000157 EDW89338.1 CH954177 EDV59529.2 NEVH01000606 PNF43431.1 AE013599 AY058326 AAF59099.4 AAL13555.1 ATLV01013334 KE524853 KFB37687.1 ACL83072.1 CH963849 EDW74227.1 GDHF01015996 JAI36318.1 CH916367 EDW00806.1 CH480816 EDW46928.1 AGBW02013359 OWR43531.1 GAMC01019594 JAB86961.1 GAKP01013697 JAC45255.1 GBXI01016298 GBXI01007032 GBXI01004348 JAC97993.1 JAD07260.1 JAD09944.1 OUUW01000001 SPP73705.1 PYGN01000167 PSN52296.1 CH933808 EDW10136.1 CH940648 EDW60839.1 JRES01000171 KNC33669.1 CM000071 KRT01637.1 KRT01636.1 AAAB01008986 EAA00363.3 CH479181 EDW31615.1 EAL24869.3 KPJ08593.1 CP012524 ALC40915.1 JXUM01049812 KQ561622 KXJ77978.1 JXUM01003156 KQ560154 KXJ84146.1 ADMH02000612 ETN65624.1 ATLV01015142 KE525003 KFB40120.1 KZ288296 PBC28930.1 KK852575 KDR21025.1 CH964291 EDW86510.1 QOIP01000009 RLU19038.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000007151
UP000053240
+ More
UP000053097 UP000078492 UP000078541 UP000037510 UP000019118 UP000078540 UP000005205 UP000078542 UP000076502 UP000242457 UP000008237 UP000075809 UP000005203 UP000002358 UP000053105 UP000053825 UP000215335 UP000007755 UP000002320 UP000069940 UP000249989 UP000008820 UP000000673 UP000069272 UP000030742 UP000075886 UP000027135 UP000075880 UP000075884 UP000076408 UP000075901 UP000075885 UP000075903 UP000075882 UP000075920 UP000075840 UP000076407 UP000007801 UP000075883 UP000192221 UP000002282 UP000008711 UP000235965 UP000000803 UP000030765 UP000075881 UP000075900 UP000007798 UP000001070 UP000001292 UP000075902 UP000268350 UP000245037 UP000009192 UP000008792 UP000037069 UP000001819 UP000007062 UP000008744 UP000092553 UP000091820 UP000095300 UP000279307
UP000053097 UP000078492 UP000078541 UP000037510 UP000019118 UP000078540 UP000005205 UP000078542 UP000076502 UP000242457 UP000008237 UP000075809 UP000005203 UP000002358 UP000053105 UP000053825 UP000215335 UP000007755 UP000002320 UP000069940 UP000249989 UP000008820 UP000000673 UP000069272 UP000030742 UP000075886 UP000027135 UP000075880 UP000075884 UP000076408 UP000075901 UP000075885 UP000075903 UP000075882 UP000075920 UP000075840 UP000076407 UP000007801 UP000075883 UP000192221 UP000002282 UP000008711 UP000235965 UP000000803 UP000030765 UP000075881 UP000075900 UP000007798 UP000001070 UP000001292 UP000075902 UP000268350 UP000245037 UP000009192 UP000008792 UP000037069 UP000001819 UP000007062 UP000008744 UP000092553 UP000091820 UP000095300 UP000279307
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9J7N2
A0A2A4IZ62
A0A2H1V795
A0A194PX02
A0A3S2M441
A0A212FBL2
+ More
A0A194QTF7 A0A1L8D8Y9 A0A026VWA6 A0A195DKK0 A0A195F7N1 A0A0C9QYB7 J3JY45 A0A0L7LV09 A0A1Y1NE52 A0A1Y1N7J2 N6TVP6 A0A195B2U2 A0A158NNG4 A0A195D7D3 A0A154PTD3 A0A2A3EKV3 E2C1A8 A0A151WWE6 A0A088A8U6 K7J0X6 A0A0M8ZVZ2 A0A0L7R3U9 A0A232ELQ6 F4X5B7 B0W824 A0A1Q3FMX6 A0A182GYB1 Q17DP2 W5JS61 A0A182FTU0 U4UDQ6 A0A182QVN0 A0A067R4X6 A0A182J1T0 A0A182MYP9 A0A182Y1N0 A0A182SA02 A0A182P6J5 A0A182VCI3 A0A182KSH7 A0A182WG56 A0A182HQ79 A0A182WZN3 B3MFZ2 A0A2A4IZE4 A0A2H1VHH4 A0A0J9R7Y3 A0A182MMG8 A0A0J9R9A0 A0A1W4UK09 B4P2S7 B3N8Q2 A0A2J7RRG4 Q7K3V9 A0A084VI93 B7YZU1 A0A182JUQ7 A0A182RTK5 B4MQ38 A0A0K8VBR6 B4J5W3 B4HRW5 A0A212EPZ6 W8B145 A0A034VTW1 A0A182TP89 A0A0A1X942 A0A3B0IZM8 A0A2P8Z727 B4KPE0 B4LLA2 A0A0L0CQF6 A0A0R3NRL5 A0A0R3NL82 Q7Q0A1 B4GDD9 Q292J3 A0A194QSV0 A0A0M4EBJ8 A0A182G9R9 A0A1A9W8X4 A0A182GWR4 A0A182FIU1 A0A1I8NQL3 W5JS93 A0A084VQ76 A0A1A9WHR3 A0A2A3EB67 A0A1I8PPG6 A0A088A7F7 A0A067RMS8 B4NPS0 A0A182KH76 A0A3L8DGP3
A0A194QTF7 A0A1L8D8Y9 A0A026VWA6 A0A195DKK0 A0A195F7N1 A0A0C9QYB7 J3JY45 A0A0L7LV09 A0A1Y1NE52 A0A1Y1N7J2 N6TVP6 A0A195B2U2 A0A158NNG4 A0A195D7D3 A0A154PTD3 A0A2A3EKV3 E2C1A8 A0A151WWE6 A0A088A8U6 K7J0X6 A0A0M8ZVZ2 A0A0L7R3U9 A0A232ELQ6 F4X5B7 B0W824 A0A1Q3FMX6 A0A182GYB1 Q17DP2 W5JS61 A0A182FTU0 U4UDQ6 A0A182QVN0 A0A067R4X6 A0A182J1T0 A0A182MYP9 A0A182Y1N0 A0A182SA02 A0A182P6J5 A0A182VCI3 A0A182KSH7 A0A182WG56 A0A182HQ79 A0A182WZN3 B3MFZ2 A0A2A4IZE4 A0A2H1VHH4 A0A0J9R7Y3 A0A182MMG8 A0A0J9R9A0 A0A1W4UK09 B4P2S7 B3N8Q2 A0A2J7RRG4 Q7K3V9 A0A084VI93 B7YZU1 A0A182JUQ7 A0A182RTK5 B4MQ38 A0A0K8VBR6 B4J5W3 B4HRW5 A0A212EPZ6 W8B145 A0A034VTW1 A0A182TP89 A0A0A1X942 A0A3B0IZM8 A0A2P8Z727 B4KPE0 B4LLA2 A0A0L0CQF6 A0A0R3NRL5 A0A0R3NL82 Q7Q0A1 B4GDD9 Q292J3 A0A194QSV0 A0A0M4EBJ8 A0A182G9R9 A0A1A9W8X4 A0A182GWR4 A0A182FIU1 A0A1I8NQL3 W5JS93 A0A084VQ76 A0A1A9WHR3 A0A2A3EB67 A0A1I8PPG6 A0A088A7F7 A0A067RMS8 B4NPS0 A0A182KH76 A0A3L8DGP3
Ontologies
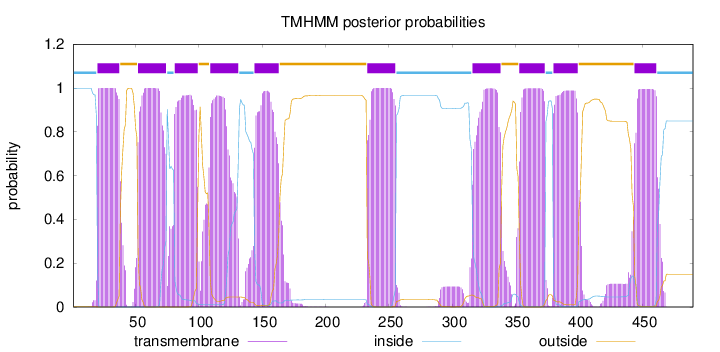
Topology
Length:
490
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
216.034
Exp number, first 60 AAs:
28.5398
Total prob of N-in:
0.99924
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 37
outside
38 - 51
TMhelix
52 - 74
inside
75 - 80
TMhelix
81 - 99
outside
100 - 108
TMhelix
109 - 131
inside
132 - 143
TMhelix
144 - 163
outside
164 - 232
TMhelix
233 - 255
inside
256 - 315
TMhelix
316 - 338
outside
339 - 352
TMhelix
353 - 373
inside
374 - 379
TMhelix
380 - 399
outside
400 - 443
TMhelix
444 - 461
inside
462 - 490
Population Genetic Test Statistics
Pi
31.520115
Theta
172.532468
Tajima's D
2.552705
CLR
0.930868
CSRT
0.949302534873256
Interpretation
Uncertain
