Pre Gene Modal
BGIBMGA003345
Annotation
PREDICTED:_UNC93-like_protein_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.975
Sequence
CDS
ATGGCGGAAGAAGTAAACAGTAAGAGAGAGGTTCAGGTGCACGGAACTGGAAGAATAGTGAAGAATGTTCTGGTGATAAGTTTGGCGTTTATGACGCAATTCACTGCGTACAGCGGTGCGGCGAATCTGCAGAGCTCGATAAACGCTAGTGCTGGCCTCGGAACCGCCGCACTGGCCGCCGTGTACGCGGGACTCATCCTCTCAAACATATTCCTGCCTGTATTTGTCATCAAGTGGCTCGGTACGAAGTGGACAATATCCTTCTCGTTTCTCACGTATATGCCGTATATAGCGGCGCAGCTGTATCCCCGATTCTACACCATGATCCCGGCGGGACTGCTCGTGGGTCTAGGCGGCGGACCTTTATGGTGTGCCAAATGCACTTATTTATACGTCATGTCTGAAGCTCATAGTGCGATTTCGAAGATAACAGCCGAAGCGTTGCTCGTACGCTTCTTAGGAATATTCTTTATGATATTTCATACGAGCCAAATTTGGGGTAACCTCATATCGTCTCTAGGAATTTATCTAGCTTGTATGGCTGGAGCTGCTATTATAGTTGCCGTTGGAGTGGATCCGATGAAAAAGTTTGATACGAGAGGCAGCGGCTCTGGTACGGGTCTGTCTGGGTTGGCATTGTTGGCCGCAACTCTAAAGCTGCTAATCGAACCGAATCAAATCTTAATGATTCCAATAAACGTCTTCTTAGGATTCCAACAAGCGTTTTTTGGGGCGGACTTTACTGCGGCCTTCGTGTCTTGTGCCGTCGGCACGGGGTCCGTTGGTTTTGTTATGATGACTTATGGATTGGCTGATGCAATAGGCTGTATCGTAAATGGATATATAGCAAAGATAATCGATCGTCTGCCTTTAATATGTATAGCTACTATAATAAACATTGCTATTTTGACGACGCTATTGACGTGGCATCCTAATGGTGGACAAGCCTATGTGCTCTACATCATAATTGTTGTTTTGGGATTCAGCGATTCGATCTGGATCGTCCAGATTAATGCGTACTATAGCATTTTGTTTCCTGGAAGAGAAGAGGCAGCTTTCTCAAACTTCAGGCTATGGGAGTCGGTCGGTTATATTGTAGCTTACGTGATCTCTCCATACCTCAGAACCAGCGTTAAGACCTACCTTATGTTATCATTACTAGTTTTAGGATTATCATGTTATTTCACTGTGGAATATCGAGAAAGAAGGAAAAAGTCGAAGTTGAATACGCCTGATAGGTACTGTTTGGAGTACAAGGCAAATATTTGTCTAGCATCTTTAGGCAAATTTGAGGGTCTTTGTCACTTAAAGTTCGTGATATAA
Protein
MAEEVNSKREVQVHGTGRIVKNVLVISLAFMTQFTAYSGAANLQSSINASAGLGTAALAAVYAGLILSNIFLPVFVIKWLGTKWTISFSFLTYMPYIAAQLYPRFYTMIPAGLLVGLGGGPLWCAKCTYLYVMSEAHSAISKITAEALLVRFLGIFFMIFHTSQIWGNLISSLGIYLACMAGAAIIVAVGVDPMKKFDTRGSGSGTGLSGLALLAATLKLLIEPNQILMIPINVFLGFQQAFFGADFTAAFVSCAVGTGSVGFVMMTYGLADAIGCIVNGYIAKIIDRLPLICIATIINIAILTTLLTWHPNGGQAYVLYIIIVVLGFSDSIWIVQINAYYSILFPGREEAAFSNFRLWESVGYIVAYVISPYLRTSVKTYLMLSLLVLGLSCYFTVEYRERRKKSKLNTPDRYCLEYKANICLASLGKFEGLCHLKFVI
Summary
Uniprot
A0A2H1VHH4
A0A2A4IZE4
A0A212EPZ6
A0A194PX02
A0A2A4IWC9
A0A067R4X6
+ More
A0A151WWE6 A0A195D7D3 A0A212EPD6 A0A195DKK0 A0A195F7N1 A0A158NNG4 F4X5B7 A0A2P8YQH7 A0A1E1WT02 A0A195B2U2 A0A2J7RRG4 A0A067RMS8 K7J0X6 A0A232ELQ6 A0A2P8Z727 A0A2A3EB67 A0A088A7F7 A0A1B6G7H6 A0A1J1IIF3 A0A034W1Q1 A0A034W050 A0A182GWR4 A0A182G9R9 A0A0C9Q4D2 K7IW01 E2C1A8 A0A139WL24 W8AD07 A0A158NFD5 A0A1Y1NB36 A0A195CTX8 A0A1B6CT84 A0A0A1XKD8 A0A0K8V7B1 A0A151JZ88 A0A0A1WEG1 A0A1B6DYS0 A0A1B6CEC5 A0A1Y1NB73 N6TTC5 Q17DS4 E9IAP3 A0A1B6M7T8 B0W7P2 A0A1Q3FR00 Q6BCZ5 A0A0L7R3U9 J9JT06 A0A2J7QTX9 A0A182MYP9 A0A182JUQ7 A0A182P6J5 A0A0M8ZVZ2 A0A182VCI3 A0A182J1T0 Q7Q0A1 A0A1B0ETI9 A0A182Y1N0 A0A182MMG8 A0A182SA02 A0A0L0CQF6 A0A182RTK5 A0A1A9WHR3 T1PC15 A0A1A9YQC3 A0A1B0DJV2 A0A1W4VWV5 A0A1D2NHD7 B4JN53 A0A084VQ76 A0A1B0CSH5 A0A2P2IDI8 A0A1B0BLH2 A0A1B0A1E3 A0A1D2MQA9 A0A1A9VWJ5 A0A1B0FCP8 A0A1B0B4K1 A0A1B0GBG4 A0A3R7NVZ3 A0A182LVV9 A0A0K2UNC0
A0A151WWE6 A0A195D7D3 A0A212EPD6 A0A195DKK0 A0A195F7N1 A0A158NNG4 F4X5B7 A0A2P8YQH7 A0A1E1WT02 A0A195B2U2 A0A2J7RRG4 A0A067RMS8 K7J0X6 A0A232ELQ6 A0A2P8Z727 A0A2A3EB67 A0A088A7F7 A0A1B6G7H6 A0A1J1IIF3 A0A034W1Q1 A0A034W050 A0A182GWR4 A0A182G9R9 A0A0C9Q4D2 K7IW01 E2C1A8 A0A139WL24 W8AD07 A0A158NFD5 A0A1Y1NB36 A0A195CTX8 A0A1B6CT84 A0A0A1XKD8 A0A0K8V7B1 A0A151JZ88 A0A0A1WEG1 A0A1B6DYS0 A0A1B6CEC5 A0A1Y1NB73 N6TTC5 Q17DS4 E9IAP3 A0A1B6M7T8 B0W7P2 A0A1Q3FR00 Q6BCZ5 A0A0L7R3U9 J9JT06 A0A2J7QTX9 A0A182MYP9 A0A182JUQ7 A0A182P6J5 A0A0M8ZVZ2 A0A182VCI3 A0A182J1T0 Q7Q0A1 A0A1B0ETI9 A0A182Y1N0 A0A182MMG8 A0A182SA02 A0A0L0CQF6 A0A182RTK5 A0A1A9WHR3 T1PC15 A0A1A9YQC3 A0A1B0DJV2 A0A1W4VWV5 A0A1D2NHD7 B4JN53 A0A084VQ76 A0A1B0CSH5 A0A2P2IDI8 A0A1B0BLH2 A0A1B0A1E3 A0A1D2MQA9 A0A1A9VWJ5 A0A1B0FCP8 A0A1B0B4K1 A0A1B0GBG4 A0A3R7NVZ3 A0A182LVV9 A0A0K2UNC0
Pubmed
EMBL
ODYU01002414
SOQ39862.1
NWSH01005059
PCG64473.1
AGBW02013359
OWR43531.1
+ More
KQ459595 KPI95670.1 NWSH01005369 PCG64267.1 KK852890 KDR14271.1 KQ982691 KYQ52224.1 KQ976749 KYN08786.1 AGBW02013486 OWR43353.1 KQ980765 KYN13376.1 KQ981744 KYN36192.1 ADTU01021539 GL888705 EGI58365.1 PYGN01000431 PSN46516.1 GDQN01004664 GDQN01000987 JAT86390.1 JAT90067.1 KQ976641 KYM78798.1 NEVH01000606 PNF43431.1 KK852575 KDR21025.1 NNAY01003515 OXU19293.1 PYGN01000167 PSN52296.1 KZ288296 PBC28930.1 GECZ01011381 JAS58388.1 CVRI01000048 CRK98846.1 GAKP01011269 JAC47683.1 GAKP01011270 JAC47682.1 JXUM01003156 KQ560154 KXJ84146.1 JXUM01049812 KQ561622 KXJ77978.1 GBYB01008918 JAG78685.1 GL451914 EFN78270.1 KQ971324 KYB28531.1 GAMC01020698 GAMC01020695 JAB85860.1 ADTU01014275 ADTU01014276 ADTU01014277 ADTU01014278 GEZM01007503 GEZM01007501 JAV95142.1 KQ977279 KYN03977.1 GEDC01020618 JAS16680.1 GBXI01002962 JAD11330.1 GDHF01017512 JAI34802.1 KQ981410 KYN41916.1 GBXI01017509 JAC96782.1 GEDC01006499 JAS30799.1 GEDC01025529 JAS11769.1 GEZM01007502 JAV95141.1 APGK01019729 APGK01019730 KB740120 ENN81323.1 CH477291 EAT44590.1 GL762047 EFZ22329.1 GEBQ01007982 JAT31995.1 DS231855 EDS38186.1 GFDL01004994 JAV30051.1 AY665388 AAT76555.1 KQ414661 KOC65552.1 ABLF02027629 ABLF02027631 NEVH01011192 PNF32038.1 KQ435855 KOX70659.1 AAAB01008986 EAA00363.3 AJWK01010483 AXCM01002861 JRES01000171 KNC33669.1 KA645473 AFP60102.1 AJVK01001114 LJIJ01000037 ODN04684.1 CH916371 EDV92146.1 ATLV01015142 KE525003 KFB40120.1 AJWK01026073 IACF01006490 LAB72073.1 JXJN01016363 LJIJ01000695 ODM95223.1 CCAG010001429 CCAG010001430 CCAG010001431 CCAG010001432 CCAG010001433 JXJN01008344 JXJN01008345 CCAG010014002 CCAG010014003 QCYY01002711 ROT68180.1 AXCM01000181 HACA01021880 CDW39241.1
KQ459595 KPI95670.1 NWSH01005369 PCG64267.1 KK852890 KDR14271.1 KQ982691 KYQ52224.1 KQ976749 KYN08786.1 AGBW02013486 OWR43353.1 KQ980765 KYN13376.1 KQ981744 KYN36192.1 ADTU01021539 GL888705 EGI58365.1 PYGN01000431 PSN46516.1 GDQN01004664 GDQN01000987 JAT86390.1 JAT90067.1 KQ976641 KYM78798.1 NEVH01000606 PNF43431.1 KK852575 KDR21025.1 NNAY01003515 OXU19293.1 PYGN01000167 PSN52296.1 KZ288296 PBC28930.1 GECZ01011381 JAS58388.1 CVRI01000048 CRK98846.1 GAKP01011269 JAC47683.1 GAKP01011270 JAC47682.1 JXUM01003156 KQ560154 KXJ84146.1 JXUM01049812 KQ561622 KXJ77978.1 GBYB01008918 JAG78685.1 GL451914 EFN78270.1 KQ971324 KYB28531.1 GAMC01020698 GAMC01020695 JAB85860.1 ADTU01014275 ADTU01014276 ADTU01014277 ADTU01014278 GEZM01007503 GEZM01007501 JAV95142.1 KQ977279 KYN03977.1 GEDC01020618 JAS16680.1 GBXI01002962 JAD11330.1 GDHF01017512 JAI34802.1 KQ981410 KYN41916.1 GBXI01017509 JAC96782.1 GEDC01006499 JAS30799.1 GEDC01025529 JAS11769.1 GEZM01007502 JAV95141.1 APGK01019729 APGK01019730 KB740120 ENN81323.1 CH477291 EAT44590.1 GL762047 EFZ22329.1 GEBQ01007982 JAT31995.1 DS231855 EDS38186.1 GFDL01004994 JAV30051.1 AY665388 AAT76555.1 KQ414661 KOC65552.1 ABLF02027629 ABLF02027631 NEVH01011192 PNF32038.1 KQ435855 KOX70659.1 AAAB01008986 EAA00363.3 AJWK01010483 AXCM01002861 JRES01000171 KNC33669.1 KA645473 AFP60102.1 AJVK01001114 LJIJ01000037 ODN04684.1 CH916371 EDV92146.1 ATLV01015142 KE525003 KFB40120.1 AJWK01026073 IACF01006490 LAB72073.1 JXJN01016363 LJIJ01000695 ODM95223.1 CCAG010001429 CCAG010001430 CCAG010001431 CCAG010001432 CCAG010001433 JXJN01008344 JXJN01008345 CCAG010014002 CCAG010014003 QCYY01002711 ROT68180.1 AXCM01000181 HACA01021880 CDW39241.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000027135
UP000075809
UP000078542
+ More
UP000078492 UP000078541 UP000005205 UP000007755 UP000245037 UP000078540 UP000235965 UP000002358 UP000215335 UP000242457 UP000005203 UP000183832 UP000069940 UP000249989 UP000008237 UP000007266 UP000019118 UP000008820 UP000002320 UP000053825 UP000007819 UP000075884 UP000075881 UP000075885 UP000053105 UP000075903 UP000075880 UP000007062 UP000092461 UP000076408 UP000075883 UP000075901 UP000037069 UP000075900 UP000091820 UP000095301 UP000092443 UP000092462 UP000192221 UP000094527 UP000001070 UP000030765 UP000092460 UP000092445 UP000078200 UP000092444 UP000283509
UP000078492 UP000078541 UP000005205 UP000007755 UP000245037 UP000078540 UP000235965 UP000002358 UP000215335 UP000242457 UP000005203 UP000183832 UP000069940 UP000249989 UP000008237 UP000007266 UP000019118 UP000008820 UP000002320 UP000053825 UP000007819 UP000075884 UP000075881 UP000075885 UP000053105 UP000075903 UP000075880 UP000007062 UP000092461 UP000076408 UP000075883 UP000075901 UP000037069 UP000075900 UP000091820 UP000095301 UP000092443 UP000092462 UP000192221 UP000094527 UP000001070 UP000030765 UP000092460 UP000092445 UP000078200 UP000092444 UP000283509
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2H1VHH4
A0A2A4IZE4
A0A212EPZ6
A0A194PX02
A0A2A4IWC9
A0A067R4X6
+ More
A0A151WWE6 A0A195D7D3 A0A212EPD6 A0A195DKK0 A0A195F7N1 A0A158NNG4 F4X5B7 A0A2P8YQH7 A0A1E1WT02 A0A195B2U2 A0A2J7RRG4 A0A067RMS8 K7J0X6 A0A232ELQ6 A0A2P8Z727 A0A2A3EB67 A0A088A7F7 A0A1B6G7H6 A0A1J1IIF3 A0A034W1Q1 A0A034W050 A0A182GWR4 A0A182G9R9 A0A0C9Q4D2 K7IW01 E2C1A8 A0A139WL24 W8AD07 A0A158NFD5 A0A1Y1NB36 A0A195CTX8 A0A1B6CT84 A0A0A1XKD8 A0A0K8V7B1 A0A151JZ88 A0A0A1WEG1 A0A1B6DYS0 A0A1B6CEC5 A0A1Y1NB73 N6TTC5 Q17DS4 E9IAP3 A0A1B6M7T8 B0W7P2 A0A1Q3FR00 Q6BCZ5 A0A0L7R3U9 J9JT06 A0A2J7QTX9 A0A182MYP9 A0A182JUQ7 A0A182P6J5 A0A0M8ZVZ2 A0A182VCI3 A0A182J1T0 Q7Q0A1 A0A1B0ETI9 A0A182Y1N0 A0A182MMG8 A0A182SA02 A0A0L0CQF6 A0A182RTK5 A0A1A9WHR3 T1PC15 A0A1A9YQC3 A0A1B0DJV2 A0A1W4VWV5 A0A1D2NHD7 B4JN53 A0A084VQ76 A0A1B0CSH5 A0A2P2IDI8 A0A1B0BLH2 A0A1B0A1E3 A0A1D2MQA9 A0A1A9VWJ5 A0A1B0FCP8 A0A1B0B4K1 A0A1B0GBG4 A0A3R7NVZ3 A0A182LVV9 A0A0K2UNC0
A0A151WWE6 A0A195D7D3 A0A212EPD6 A0A195DKK0 A0A195F7N1 A0A158NNG4 F4X5B7 A0A2P8YQH7 A0A1E1WT02 A0A195B2U2 A0A2J7RRG4 A0A067RMS8 K7J0X6 A0A232ELQ6 A0A2P8Z727 A0A2A3EB67 A0A088A7F7 A0A1B6G7H6 A0A1J1IIF3 A0A034W1Q1 A0A034W050 A0A182GWR4 A0A182G9R9 A0A0C9Q4D2 K7IW01 E2C1A8 A0A139WL24 W8AD07 A0A158NFD5 A0A1Y1NB36 A0A195CTX8 A0A1B6CT84 A0A0A1XKD8 A0A0K8V7B1 A0A151JZ88 A0A0A1WEG1 A0A1B6DYS0 A0A1B6CEC5 A0A1Y1NB73 N6TTC5 Q17DS4 E9IAP3 A0A1B6M7T8 B0W7P2 A0A1Q3FR00 Q6BCZ5 A0A0L7R3U9 J9JT06 A0A2J7QTX9 A0A182MYP9 A0A182JUQ7 A0A182P6J5 A0A0M8ZVZ2 A0A182VCI3 A0A182J1T0 Q7Q0A1 A0A1B0ETI9 A0A182Y1N0 A0A182MMG8 A0A182SA02 A0A0L0CQF6 A0A182RTK5 A0A1A9WHR3 T1PC15 A0A1A9YQC3 A0A1B0DJV2 A0A1W4VWV5 A0A1D2NHD7 B4JN53 A0A084VQ76 A0A1B0CSH5 A0A2P2IDI8 A0A1B0BLH2 A0A1B0A1E3 A0A1D2MQA9 A0A1A9VWJ5 A0A1B0FCP8 A0A1B0B4K1 A0A1B0GBG4 A0A3R7NVZ3 A0A182LVV9 A0A0K2UNC0
Ontologies
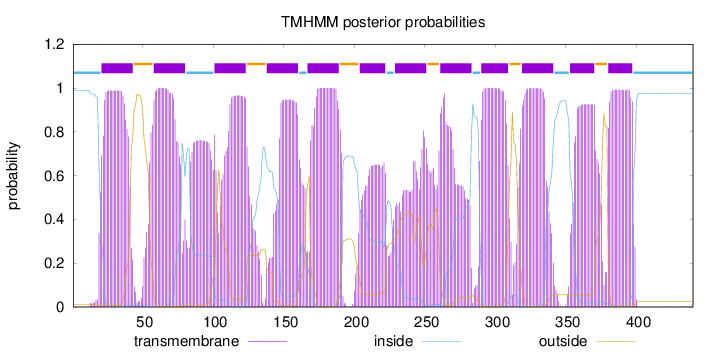
Topology
Length:
440
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
249.27189
Exp number, first 60 AAs:
27.4453
Total prob of N-in:
0.98958
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 57
TMhelix
58 - 80
inside
81 - 100
TMhelix
101 - 123
outside
124 - 137
TMhelix
138 - 160
inside
161 - 166
TMhelix
167 - 189
outside
190 - 203
TMhelix
204 - 222
inside
223 - 228
TMhelix
229 - 251
outside
252 - 260
TMhelix
261 - 283
inside
284 - 289
TMhelix
290 - 309
outside
310 - 318
TMhelix
319 - 341
inside
342 - 352
TMhelix
353 - 370
outside
371 - 379
TMhelix
380 - 397
inside
398 - 440
Population Genetic Test Statistics
Pi
386.465265
Theta
186.525271
Tajima's D
3.519161
CLR
0.38946
CSRT
0.994000299985001
Interpretation
Uncertain
