Pre Gene Modal
BGIBMGA005286
Annotation
RNA_m5u_methyltransferase_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 1.832
Sequence
CDS
ATGGATAAATCAAATCGCAGTGTTATTGAAGCTAACAGGAAAATATACAACGGTCTGAGTTTTGAACTAAAGCCCATTGAAAAATCTCCTGTCTTAGATGGCTACAGGAATAAATGCGAGTTTACAATTGGTATTGATGATGAGACTAAGTTACCCACCGTTGGGTTTAGATTGGGTAGTTATGTGACTGGTACAGTGGCTGTGGCACCCATAGAAATGTTGATACATATACCCGAGAGAACGAAACAGGCTGTTTTGGTATTCCAAGACTTTGTCCGTAAATCTGGATTAGCTCCATTCTCTCCGGCTGACTATTCTGGATACTGGCGTTATTTGACCGTCAGGCAGTCAACATCGAATGGTGACGTCATGTTAATTGCGTCCATGTATCCTCAGAATTTAACTGAGAGTGAAATAGAAAAAATTAAAAATGACTTGATAGAACATTGTAGTTCTAAAGAAGTTGATTGTGGGATTAAATCATTGTATTTTGAGTTGATCACTAAAAGGAGGGTTGGTGAAGAACCAACAAAGCCTACACACTTAATGGGATCAACTCATATCATTGACACAATATTGGGGAGACAGTTTCGGATTTCCCCAGAAGCATTCTTTCAGATAAACACAGCCGGTGCTGAAATACTGTATCAAAAGGCTATTGAACTGAGTGAAGTCAAAGAGGATTCGACTGTGATAGATATTTGTTGTGGGACCGGCACGATAGGCCTGTGTTTTGCTAAACACTGCGGTAAAGTTTTAGGCTTAGAATTGGTACCGGAAGCTGTGAAGGACGCTAAAGCAAATGCTGAACTTAACGAAATCGAAAATAGTGAGTTCTACACGGGGAAAGCTGAAGATATACTACCCTCGGTGCTGGCCAGGGCCACCGGAGACGATGTTATTGCGATTGTTGATCCCCCAAGAGCTGGACTCCACATGCGCGCCGTGACGCAGCTGCGCAACAGCCGGCGCGTGCGGCGCGTGCTGTACGTGTCGTGCCGGCCCGCCGCCGCGCTCAAGAACTGGGTGGACCTCGCTCGGCCCTCCTCCAAGACTCTCCGCGGTGCCCCCTTCCGCCCGCGCCGGGCCCTGCCCCTCGACATGTTCCCGCACACCGACCACATCGAGCTCGCCTTGCTGTTCGAGAGGGAACAGACGAAAGAAGATACCGAAGGAAGTTCGGAAACAGTGGAAGACAGCAGTGCGGCACCGGACGTGGAGAATAAAGATAATAATTAG
Protein
MDKSNRSVIEANRKIYNGLSFELKPIEKSPVLDGYRNKCEFTIGIDDETKLPTVGFRLGSYVTGTVAVAPIEMLIHIPERTKQAVLVFQDFVRKSGLAPFSPADYSGYWRYLTVRQSTSNGDVMLIASMYPQNLTESEIEKIKNDLIEHCSSKEVDCGIKSLYFELITKRRVGEEPTKPTHLMGSTHIIDTILGRQFRISPEAFFQINTAGAEILYQKAIELSEVKEDSTVIDICCGTGTIGLCFAKHCGKVLGLELVPEAVKDAKANAELNEIENSEFYTGKAEDILPSVLARATGDDVIAIVDPPRAGLHMRAVTQLRNSRRVRRVLYVSCRPAAALKNWVDLARPSSKTLRGAPFRPRRALPLDMFPHTDHIELALLFEREQTKEDTEGSSETVEDSSAAPDVENKDNN
Summary
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. RNA M5U methyltransferase family.
Uniprot
H9J6Z4
A0A2H1WMK9
A0A2H1UZQ0
A0A2A4IVR4
A0A212EIN2
A0A194PQS9
+ More
A0A2Z5U7M9 S4PLB7 A0A2J7PM06 A0A067R8Z7 A0A2J7PM02 Q17NM7 A0A182GFX9 D6WVM0 A0A1Y1LUI3 A0A026VSE5 A0A3L8DCG6 A0A1Y1LUL4 W5JGM8 A0A1Q3EZ50 B0X3M6 A0A151WWA3 F4X6B1 A0A151J3R3 A0A0J7KYM6 A0A158P0Q6 A0A084WIW9 A0A195FYY7 A0A195B1U1 A0A2M4AVF9 E2AVU8 A0A3F2YXP0 A0A195CXF2 N6U7G0 B0WFQ3 A0A182JJ72 A0A182SF43 A0A3F2YX70 A0A1B6D834 E2CA93 A0A182M023 Q7Q0E4 A0A182KSA4 A0A3F2YU11 A0A1Y9IVJ4 A0A182UHR1 A0A182V9X5 E9IYJ1 E0VCT9 A0A1L8DC01 B4LD33 A0A1I8NGG8 A0A154PSV8 T1PCJ6 A0A1B6F487 A0A310S4M4 Q29DJ3 A0A3B0J8E6 A0A0L7QVJ7 B4N4R7 Q95R49 B4PH07 A0A1W4UBW2 Q9VVV7 B4IFY7 A0A0J9UP18 B5RIS0 B4QQB8 B4KZ91 B4IU84 W8C554 B3NDW6 A0A1B6J213 A0A2A3ELA1 A0A182Y1G5 B4IY53 A0A0A1XQN7 B3M7Q0 A0A1I8NZX1 A0A0M4EJK6 A0A1B6MKH0 A0A087ZWF9 A0A1A9W3F1 A0A034WPP2 A0A0L0CAS5 A0A0N1IU26 A0A0K8W5B7 A0A1J1IL58 A0A232FJD1 K7IZ01 A0A2J7PK04 A0A0V0G615 A0A069DVB8 A0A087SY41 A0A023F1R7 C3XVW9 A0A0P4WAU1 A0A146KMC2 K1QUQ4 A0A1A9YID1
A0A2Z5U7M9 S4PLB7 A0A2J7PM06 A0A067R8Z7 A0A2J7PM02 Q17NM7 A0A182GFX9 D6WVM0 A0A1Y1LUI3 A0A026VSE5 A0A3L8DCG6 A0A1Y1LUL4 W5JGM8 A0A1Q3EZ50 B0X3M6 A0A151WWA3 F4X6B1 A0A151J3R3 A0A0J7KYM6 A0A158P0Q6 A0A084WIW9 A0A195FYY7 A0A195B1U1 A0A2M4AVF9 E2AVU8 A0A3F2YXP0 A0A195CXF2 N6U7G0 B0WFQ3 A0A182JJ72 A0A182SF43 A0A3F2YX70 A0A1B6D834 E2CA93 A0A182M023 Q7Q0E4 A0A182KSA4 A0A3F2YU11 A0A1Y9IVJ4 A0A182UHR1 A0A182V9X5 E9IYJ1 E0VCT9 A0A1L8DC01 B4LD33 A0A1I8NGG8 A0A154PSV8 T1PCJ6 A0A1B6F487 A0A310S4M4 Q29DJ3 A0A3B0J8E6 A0A0L7QVJ7 B4N4R7 Q95R49 B4PH07 A0A1W4UBW2 Q9VVV7 B4IFY7 A0A0J9UP18 B5RIS0 B4QQB8 B4KZ91 B4IU84 W8C554 B3NDW6 A0A1B6J213 A0A2A3ELA1 A0A182Y1G5 B4IY53 A0A0A1XQN7 B3M7Q0 A0A1I8NZX1 A0A0M4EJK6 A0A1B6MKH0 A0A087ZWF9 A0A1A9W3F1 A0A034WPP2 A0A0L0CAS5 A0A0N1IU26 A0A0K8W5B7 A0A1J1IL58 A0A232FJD1 K7IZ01 A0A2J7PK04 A0A0V0G615 A0A069DVB8 A0A087SY41 A0A023F1R7 C3XVW9 A0A0P4WAU1 A0A146KMC2 K1QUQ4 A0A1A9YID1
Pubmed
19121390
22118469
26354079
23622113
24845553
17510324
+ More
26483478 18362917 19820115 28004739 24508170 30249741 20920257 23761445 21719571 21347285 24438588 20798317 23537049 12364791 20966253 21282665 20566863 17994087 25315136 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 24495485 25244985 25830018 18057021 25348373 26108605 28648823 20075255 26334808 25474469 18563158 26823975 22992520
26483478 18362917 19820115 28004739 24508170 30249741 20920257 23761445 21719571 21347285 24438588 20798317 23537049 12364791 20966253 21282665 20566863 17994087 25315136 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 24495485 25244985 25830018 18057021 25348373 26108605 28648823 20075255 26334808 25474469 18563158 26823975 22992520
EMBL
BABH01014074
ODYU01009636
SOQ54206.1
ODYU01000020
SOQ34077.1
NWSH01005952
+ More
PCG63811.1 AGBW02014614 OWR41335.1 KQ459595 KPI95667.1 AP017551 BBB06976.1 GAIX01004105 JAA88455.1 NEVH01024421 PNF17366.1 KK852807 KDR16066.1 PNF17365.1 CH477198 EAT48287.1 JXUM01060640 KQ562111 KXJ76670.1 KQ971357 EFA08275.1 GEZM01046353 JAV77304.1 KK108837 EZA46612.1 QOIP01000010 RLU17608.1 GEZM01046349 JAV77309.1 ADMH02001279 ETN63241.1 GFDL01014527 JAV20518.1 DS232318 EDS39918.1 KQ982691 KYQ52037.1 GL888802 EGI58019.1 KQ980254 KYN17079.1 LBMM01001880 KMQ95617.1 ADTU01005733 ATLV01023952 KE525347 KFB50163.1 KQ981200 KYN45039.1 KQ976667 KYM78255.1 GGFK01011439 MBW44760.1 GL443213 EFN62370.1 KQ977141 KYN05343.1 APGK01040122 KB740975 KB632267 ENN76561.1 ERL91006.1 DS231920 EDS26397.1 AXCN02001026 GEDC01015450 JAS21848.1 GL453952 EFN75142.1 AXCM01006744 AAAB01008986 EAA00138.4 GL767018 EFZ14341.1 DS235065 EEB11195.1 GFDF01010179 JAV03905.1 CH940647 EDW69914.1 KQ435127 KZC14847.1 KA645673 AFP60302.1 GECZ01024697 JAS45072.1 KQ776210 OAD52212.1 CH379070 EAL30421.1 OUUW01000002 SPP76543.1 KQ414727 KOC62531.1 CH964095 EDW79141.2 AY061614 AAL29162.1 CM000159 EDW95377.1 AE014296 AAF49200.3 CH480834 EDW46574.1 CM002912 KMZ00486.1 BT044194 ACH92259.1 CM000363 EDX11024.1 CH933809 EDW18917.1 CH891818 EDW99947.1 GAMC01001962 JAC04594.1 CH954178 EDV52320.1 GECU01014499 JAS93207.1 KZ288215 PBC32498.1 CH916366 EDV97596.1 GBXI01000986 JAD13306.1 CH902618 EDV40978.1 KPU78867.1 CP012525 ALC45152.1 GEBQ01003566 JAT36411.1 GAKP01001381 JAC57571.1 JRES01000760 KNC28559.1 KQ435713 KOX79321.1 GDHF01006219 JAI46095.1 CVRI01000054 CRL00292.1 NNAY01000120 OXU30785.1 NEVH01024944 PNF16663.1 GECL01002687 JAP03437.1 GBGD01000861 JAC88028.1 KK112487 KFM57780.1 GBBI01003449 JAC15263.1 GG666469 EEN67912.1 GDRN01077962 JAI62655.1 GDHC01020938 JAP97690.1 JH817760 EKC32655.1
PCG63811.1 AGBW02014614 OWR41335.1 KQ459595 KPI95667.1 AP017551 BBB06976.1 GAIX01004105 JAA88455.1 NEVH01024421 PNF17366.1 KK852807 KDR16066.1 PNF17365.1 CH477198 EAT48287.1 JXUM01060640 KQ562111 KXJ76670.1 KQ971357 EFA08275.1 GEZM01046353 JAV77304.1 KK108837 EZA46612.1 QOIP01000010 RLU17608.1 GEZM01046349 JAV77309.1 ADMH02001279 ETN63241.1 GFDL01014527 JAV20518.1 DS232318 EDS39918.1 KQ982691 KYQ52037.1 GL888802 EGI58019.1 KQ980254 KYN17079.1 LBMM01001880 KMQ95617.1 ADTU01005733 ATLV01023952 KE525347 KFB50163.1 KQ981200 KYN45039.1 KQ976667 KYM78255.1 GGFK01011439 MBW44760.1 GL443213 EFN62370.1 KQ977141 KYN05343.1 APGK01040122 KB740975 KB632267 ENN76561.1 ERL91006.1 DS231920 EDS26397.1 AXCN02001026 GEDC01015450 JAS21848.1 GL453952 EFN75142.1 AXCM01006744 AAAB01008986 EAA00138.4 GL767018 EFZ14341.1 DS235065 EEB11195.1 GFDF01010179 JAV03905.1 CH940647 EDW69914.1 KQ435127 KZC14847.1 KA645673 AFP60302.1 GECZ01024697 JAS45072.1 KQ776210 OAD52212.1 CH379070 EAL30421.1 OUUW01000002 SPP76543.1 KQ414727 KOC62531.1 CH964095 EDW79141.2 AY061614 AAL29162.1 CM000159 EDW95377.1 AE014296 AAF49200.3 CH480834 EDW46574.1 CM002912 KMZ00486.1 BT044194 ACH92259.1 CM000363 EDX11024.1 CH933809 EDW18917.1 CH891818 EDW99947.1 GAMC01001962 JAC04594.1 CH954178 EDV52320.1 GECU01014499 JAS93207.1 KZ288215 PBC32498.1 CH916366 EDV97596.1 GBXI01000986 JAD13306.1 CH902618 EDV40978.1 KPU78867.1 CP012525 ALC45152.1 GEBQ01003566 JAT36411.1 GAKP01001381 JAC57571.1 JRES01000760 KNC28559.1 KQ435713 KOX79321.1 GDHF01006219 JAI46095.1 CVRI01000054 CRL00292.1 NNAY01000120 OXU30785.1 NEVH01024944 PNF16663.1 GECL01002687 JAP03437.1 GBGD01000861 JAC88028.1 KK112487 KFM57780.1 GBBI01003449 JAC15263.1 GG666469 EEN67912.1 GDRN01077962 JAI62655.1 GDHC01020938 JAP97690.1 JH817760 EKC32655.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000235965
UP000027135
+ More
UP000008820 UP000069940 UP000249989 UP000007266 UP000053097 UP000279307 UP000000673 UP000002320 UP000075809 UP000007755 UP000078492 UP000036403 UP000005205 UP000030765 UP000078541 UP000078540 UP000000311 UP000075900 UP000078542 UP000019118 UP000030742 UP000075880 UP000075901 UP000075886 UP000008237 UP000075883 UP000007062 UP000075882 UP000075881 UP000075920 UP000075902 UP000075903 UP000009046 UP000008792 UP000095301 UP000076502 UP000001819 UP000268350 UP000053825 UP000007798 UP000002282 UP000192221 UP000000803 UP000001292 UP000000304 UP000009192 UP000008711 UP000242457 UP000076408 UP000001070 UP000007801 UP000095300 UP000092553 UP000005203 UP000091820 UP000037069 UP000053105 UP000183832 UP000215335 UP000002358 UP000054359 UP000001554 UP000005408 UP000092443
UP000008820 UP000069940 UP000249989 UP000007266 UP000053097 UP000279307 UP000000673 UP000002320 UP000075809 UP000007755 UP000078492 UP000036403 UP000005205 UP000030765 UP000078541 UP000078540 UP000000311 UP000075900 UP000078542 UP000019118 UP000030742 UP000075880 UP000075901 UP000075886 UP000008237 UP000075883 UP000007062 UP000075882 UP000075881 UP000075920 UP000075902 UP000075903 UP000009046 UP000008792 UP000095301 UP000076502 UP000001819 UP000268350 UP000053825 UP000007798 UP000002282 UP000192221 UP000000803 UP000001292 UP000000304 UP000009192 UP000008711 UP000242457 UP000076408 UP000001070 UP000007801 UP000095300 UP000092553 UP000005203 UP000091820 UP000037069 UP000053105 UP000183832 UP000215335 UP000002358 UP000054359 UP000001554 UP000005408 UP000092443
Interpro
Gene 3D
ProteinModelPortal
H9J6Z4
A0A2H1WMK9
A0A2H1UZQ0
A0A2A4IVR4
A0A212EIN2
A0A194PQS9
+ More
A0A2Z5U7M9 S4PLB7 A0A2J7PM06 A0A067R8Z7 A0A2J7PM02 Q17NM7 A0A182GFX9 D6WVM0 A0A1Y1LUI3 A0A026VSE5 A0A3L8DCG6 A0A1Y1LUL4 W5JGM8 A0A1Q3EZ50 B0X3M6 A0A151WWA3 F4X6B1 A0A151J3R3 A0A0J7KYM6 A0A158P0Q6 A0A084WIW9 A0A195FYY7 A0A195B1U1 A0A2M4AVF9 E2AVU8 A0A3F2YXP0 A0A195CXF2 N6U7G0 B0WFQ3 A0A182JJ72 A0A182SF43 A0A3F2YX70 A0A1B6D834 E2CA93 A0A182M023 Q7Q0E4 A0A182KSA4 A0A3F2YU11 A0A1Y9IVJ4 A0A182UHR1 A0A182V9X5 E9IYJ1 E0VCT9 A0A1L8DC01 B4LD33 A0A1I8NGG8 A0A154PSV8 T1PCJ6 A0A1B6F487 A0A310S4M4 Q29DJ3 A0A3B0J8E6 A0A0L7QVJ7 B4N4R7 Q95R49 B4PH07 A0A1W4UBW2 Q9VVV7 B4IFY7 A0A0J9UP18 B5RIS0 B4QQB8 B4KZ91 B4IU84 W8C554 B3NDW6 A0A1B6J213 A0A2A3ELA1 A0A182Y1G5 B4IY53 A0A0A1XQN7 B3M7Q0 A0A1I8NZX1 A0A0M4EJK6 A0A1B6MKH0 A0A087ZWF9 A0A1A9W3F1 A0A034WPP2 A0A0L0CAS5 A0A0N1IU26 A0A0K8W5B7 A0A1J1IL58 A0A232FJD1 K7IZ01 A0A2J7PK04 A0A0V0G615 A0A069DVB8 A0A087SY41 A0A023F1R7 C3XVW9 A0A0P4WAU1 A0A146KMC2 K1QUQ4 A0A1A9YID1
A0A2Z5U7M9 S4PLB7 A0A2J7PM06 A0A067R8Z7 A0A2J7PM02 Q17NM7 A0A182GFX9 D6WVM0 A0A1Y1LUI3 A0A026VSE5 A0A3L8DCG6 A0A1Y1LUL4 W5JGM8 A0A1Q3EZ50 B0X3M6 A0A151WWA3 F4X6B1 A0A151J3R3 A0A0J7KYM6 A0A158P0Q6 A0A084WIW9 A0A195FYY7 A0A195B1U1 A0A2M4AVF9 E2AVU8 A0A3F2YXP0 A0A195CXF2 N6U7G0 B0WFQ3 A0A182JJ72 A0A182SF43 A0A3F2YX70 A0A1B6D834 E2CA93 A0A182M023 Q7Q0E4 A0A182KSA4 A0A3F2YU11 A0A1Y9IVJ4 A0A182UHR1 A0A182V9X5 E9IYJ1 E0VCT9 A0A1L8DC01 B4LD33 A0A1I8NGG8 A0A154PSV8 T1PCJ6 A0A1B6F487 A0A310S4M4 Q29DJ3 A0A3B0J8E6 A0A0L7QVJ7 B4N4R7 Q95R49 B4PH07 A0A1W4UBW2 Q9VVV7 B4IFY7 A0A0J9UP18 B5RIS0 B4QQB8 B4KZ91 B4IU84 W8C554 B3NDW6 A0A1B6J213 A0A2A3ELA1 A0A182Y1G5 B4IY53 A0A0A1XQN7 B3M7Q0 A0A1I8NZX1 A0A0M4EJK6 A0A1B6MKH0 A0A087ZWF9 A0A1A9W3F1 A0A034WPP2 A0A0L0CAS5 A0A0N1IU26 A0A0K8W5B7 A0A1J1IL58 A0A232FJD1 K7IZ01 A0A2J7PK04 A0A0V0G615 A0A069DVB8 A0A087SY41 A0A023F1R7 C3XVW9 A0A0P4WAU1 A0A146KMC2 K1QUQ4 A0A1A9YID1
PDB
5XJ1
E-value=3.15462e-35,
Score=372
Ontologies
GO
Topology
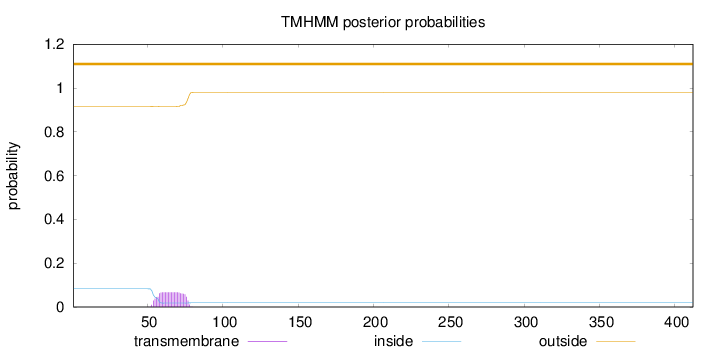
Length:
412
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.43464
Exp number, first 60 AAs:
0.36617
Total prob of N-in:
0.08349
outside
1 - 412
Population Genetic Test Statistics
Pi
281.366962
Theta
198.172031
Tajima's D
1.351028
CLR
0.013472
CSRT
0.749962501874906
Interpretation
Uncertain
