Pre Gene Modal
BGIBMGA005287
Annotation
tyramine_beta_hydroxylase_precursor_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.036
Sequence
CDS
ATGTCGAATTGGTTTTTTGTGTTTTTTCTGTCTGTTTATACATTTATTTATACGTTTTCTGTGAACGTAAGCCCAGGTCATCGTTTCGCTGAAAGTGAATCAGTGTTGGATCCAAGCGGCGAGTTTTTATTAAAGTGGCGTGTTGATTATGCCGTTCGAAAGATTAAATTTACTTTGACTGTTTCTGAAAAAGCGCCGGCCTTCAATTGGTTTGCATTAGGCTTCTCCGATCGGGGCCAACTGAATAATTCGGATGTTTGTCTGTTTTGGACAGATTATAAAGCTCGTGATCATTTCGAGGATATGCACACAAACGGACAGGGGAATTTGATACGTGATCAAAAACAAAACTGTGAAGGGTTTTATTTGAATACAAATAGCCAAAGCATAATCTTTGATCGTTTATTTGATACTTGCGATGATGATGATTACGTGATTGAGGACGGCACAGTCCATGTGGTGTGGGCTCACGGAATAGACAAACTGTTCTCATCAAAGGGCTTGTGCCTTACTTGCACCGTACCTCAGAGGCACGGCTTTGTTAGAGTAAGGCTACTAACACCTCCCGGCTTGCAAAAAGCGAATGGTTACCAATTACGAATAACAAACAAAGATCTAAAAGTACCGGGAGACGACACGACGTACTGGTGCAAAGTCGTTAGGCTGCCCGAATTCGTCACGTCAAAAGTTCATCATATAGTTCAGTTCGAATCGACAATAACTCCCGGTAACGAAGGCCTAGTTCATCACATGGAAGTCTTCTATTGCGACGAAGATCCCCACAAGGAAATGCCGGCATACGAGGGAAATTGTTTTGCCGCAGAGAGACCGGCTATTACGAAAAGTTGTTCTAAGGTAAAAGCGGCATGGGCAATGGGCGCTCCCCCTTTTACCTACCCAAAAGAAGCGGGCCTCCCACTCGGTGGCCCGAAAGCCAACAAATACGTCATGCTAGAAGTCCACTACAACAATCCAGAATTAAGGAAAGATTGGGTCGACAGTTCGGGTATTGTTCTCTATATAACTGGAAGGAAGCGTACGTACGATGCCGCCATTATGGAATTGGGCTTAGAATACACCGACAAAATGGCTATACCTGGTGAACAAAAAGCATTTCCGTTAACTGGTTACTGTATCCCTCAGTGTACTGGAGTGGGCCTACCGGATGAAGGCATTACAGTTTTTGGAAGTCAACTTCACACGCATTTAACTGGTGCCGCGGTGTGGACCAGACACTCGAGACAGGGTGTAGAGCTTCCCGTATTAAACAAAGACATGCATTATTCTACGCATTTTCAGGAAATTAGAATACTGCATCGGCCCGTTAAAGTGCTACCAGGCGACTTCTTGGAAACCACCTGCATTTACAATACGGAAGATAAGTTAAACGCGACGATCGGCGGCCACGCTATAACGGACGAAATGTGCGTTAATTACATACATTACTATCCGGCGACCGAACTCGAGGTTTGCAAGAGCGCCGTGTCGAACGAAGCACTCGAGAAATATTTCGATTTCGAAAAACGATGGGACGATATACCGATTTCGTCGAAGGCGACGCCACGTGATAATTATTTAGCGATCCAACCGTGGACTAGATTAAGAGCCGACACACTACATACGCTGTACGTCGAGTCCCCAATATCAATGCAATGCAACAAATCCGATGGGAGTCGTTTTCAAGGTGATTGGGAAGGAATTCCGATACCGAAAATAAAACTGCCGCTTTCCGAAGAAATTCGAATCTGTCCTAAAATTAACTATTTAGTCGAGACCTAA
Protein
MSNWFFVFFLSVYTFIYTFSVNVSPGHRFAESESVLDPSGEFLLKWRVDYAVRKIKFTLTVSEKAPAFNWFALGFSDRGQLNNSDVCLFWTDYKARDHFEDMHTNGQGNLIRDQKQNCEGFYLNTNSQSIIFDRLFDTCDDDDYVIEDGTVHVVWAHGIDKLFSSKGLCLTCTVPQRHGFVRVRLLTPPGLQKANGYQLRITNKDLKVPGDDTTYWCKVVRLPEFVTSKVHHIVQFESTITPGNEGLVHHMEVFYCDEDPHKEMPAYEGNCFAAERPAITKSCSKVKAAWAMGAPPFTYPKEAGLPLGGPKANKYVMLEVHYNNPELRKDWVDSSGIVLYITGRKRTYDAAIMELGLEYTDKMAIPGEQKAFPLTGYCIPQCTGVGLPDEGITVFGSQLHTHLTGAAVWTRHSRQGVELPVLNKDMHYSTHFQEIRILHRPVKVLPGDFLETTCIYNTEDKLNATIGGHAITDEMCVNYIHYYPATELEVCKSAVSNEALEKYFDFEKRWDDIPISSKATPRDNYLAIQPWTRLRADTLHTLYVESPISMQCNKSDGSRFQGDWEGIPIPKIKLPLSEEIRICPKINYLVET
Summary
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. RNA M5U methyltransferase family.
Uniprot
F1T2L6
A0A2A4IVW4
A0A194PQE1
A0A0G3VI00
A0A194QUD3
A0A212EIM8
+ More
A0A0L7LUG0 A0A1B6KW36 A0A1B6EFZ9 A0A1B6CBY6 I7C6J7 I7BWB2 I7D3M3 I7CTE1 A0A2J7PJ22 A0A067QTZ3 A0A2H8TGU2 A0A0P6H9G9 A0A1W4WN56 A0A0P6GQ74 A0A0P5LMW4 A0A0P5H9I3 A0A0P5LRQ0 A0A0P5MBJ7 W8VKK1 A0A0P5M143 A0A0P5P5B5 A0A0P6FVD3 A0A0P5H0D0 A0A0P5QZD0 A0A0P6EIS1 A0A0P5L6R5 A0A0P5NUR4 A0A0P5NQ72 A0A0P5LG82 A0A0P6EVL1 A0A0P5MNP2 A0A0P5MER8 A0A0P5M937 A0A0P5PMI4 A0A0P5PSW4 A0A0P5R021 A0A0N8DWZ2 A0A0P5MT95 A0A0P6HCP7 E0VHU4 A0A0P6ACI3 A0A0P5HAF7 A0A0P5MGX2 A0A0P5KR28 A0A0P6EZ15 A0A0P5EWS2 A0A164VW34 A0A0P6F255 A0A0P5IQN1 A0A0P5F328 D6WEK9 A0A2P8YLP8 A0A2S2N8E7 X2KVP5 J9K380 A0A0P5JYZ3 A0A0P5BG39 A0A0P6FB14 A0A0P5YSU2 A0A0P5DFR6 A0A0P5IC69 E9HK72 A0A0P5LGM6 A0A2A3EII7 A0A0P6EWW9 A0A087TQF1 W8VTH9 K7J3J6 E2B3J5 A0A0L7R8A1 A0A195FM60 A0A0P5MYZ9 A0A154PNF6 A0A151J6J3 A0A2L2YJK8 A0A195BW72 A0A158NEC4 A0A151WVS2 A0A195CCB6 A0A0C9R1Y8 A0A0P5JW18 L7LXT4 A0A232FDF1 A0A0M8ZPV4 E1ZZB1 F4WJ90 E9J1G9 T1K603 A0A1B6I325 A0A139WM35 Q147S1 A0A0P4YDU6
A0A0L7LUG0 A0A1B6KW36 A0A1B6EFZ9 A0A1B6CBY6 I7C6J7 I7BWB2 I7D3M3 I7CTE1 A0A2J7PJ22 A0A067QTZ3 A0A2H8TGU2 A0A0P6H9G9 A0A1W4WN56 A0A0P6GQ74 A0A0P5LMW4 A0A0P5H9I3 A0A0P5LRQ0 A0A0P5MBJ7 W8VKK1 A0A0P5M143 A0A0P5P5B5 A0A0P6FVD3 A0A0P5H0D0 A0A0P5QZD0 A0A0P6EIS1 A0A0P5L6R5 A0A0P5NUR4 A0A0P5NQ72 A0A0P5LG82 A0A0P6EVL1 A0A0P5MNP2 A0A0P5MER8 A0A0P5M937 A0A0P5PMI4 A0A0P5PSW4 A0A0P5R021 A0A0N8DWZ2 A0A0P5MT95 A0A0P6HCP7 E0VHU4 A0A0P6ACI3 A0A0P5HAF7 A0A0P5MGX2 A0A0P5KR28 A0A0P6EZ15 A0A0P5EWS2 A0A164VW34 A0A0P6F255 A0A0P5IQN1 A0A0P5F328 D6WEK9 A0A2P8YLP8 A0A2S2N8E7 X2KVP5 J9K380 A0A0P5JYZ3 A0A0P5BG39 A0A0P6FB14 A0A0P5YSU2 A0A0P5DFR6 A0A0P5IC69 E9HK72 A0A0P5LGM6 A0A2A3EII7 A0A0P6EWW9 A0A087TQF1 W8VTH9 K7J3J6 E2B3J5 A0A0L7R8A1 A0A195FM60 A0A0P5MYZ9 A0A154PNF6 A0A151J6J3 A0A2L2YJK8 A0A195BW72 A0A158NEC4 A0A151WVS2 A0A195CCB6 A0A0C9R1Y8 A0A0P5JW18 L7LXT4 A0A232FDF1 A0A0M8ZPV4 E1ZZB1 F4WJ90 E9J1G9 T1K603 A0A1B6I325 A0A139WM35 Q147S1 A0A0P4YDU6
Pubmed
EMBL
AB621950
BAK09201.1
NWSH01005952
PCG63809.1
KQ459595
KPI95666.1
+ More
KP657630 AKL78855.1 KQ461154 KPJ08590.1 AGBW02014614 OWR41334.1 JTDY01000066 KOB79082.1 GEBQ01024543 JAT15434.1 GEDC01000504 JAS36794.1 GEDC01026310 JAS10988.1 JQ316454 AFO63077.1 JQ316456 AFO63079.1 JQ316457 AFO63080.1 JQ316453 JQ316455 AFO63076.1 AFO63078.1 NEVH01024952 PNF16319.1 KK853274 KDR09110.1 GFXV01001510 MBW13315.1 GDIQ01023602 JAN71135.1 GDIQ01097519 GDIQ01095784 GDIQ01031867 JAN62870.1 GDIQ01178047 JAK73678.1 GDIQ01236230 JAK15495.1 GDIQ01176392 JAK75333.1 GDIQ01160609 JAK91116.1 AB897808 BAO52001.1 GDIQ01162070 JAK89655.1 GDIQ01154421 JAK97304.1 GDIQ01060043 JAN34694.1 GDIQ01235080 JAK16645.1 GDIQ01127815 JAL23911.1 GDIQ01062186 JAN32551.1 GDIQ01179489 JAK72236.1 GDIQ01137464 JAL14262.1 GDIQ01141812 JAL09914.1 GDIQ01180900 GDIQ01067901 JAK70825.1 GDIQ01174142 GDIQ01057408 JAN37329.1 GDIQ01156905 JAK94820.1 GDIQ01160608 JAK91117.1 GDIQ01158906 JAK92819.1 GDIQ01131760 GDIQ01130468 JAL19966.1 GDIQ01144857 GDIQ01082337 GDIQ01042848 JAL06869.1 GDIQ01107474 JAL44252.1 GDIQ01083732 JAN11005.1 GDIQ01151526 JAL00200.1 GDIQ01030623 JAN64114.1 DS235171 EEB12867.1 GDIP01032747 JAM70968.1 GDIQ01229285 JAK22440.1 GDIQ01179490 JAK72235.1 GDIQ01180901 JAK70824.1 GDIQ01063606 JAN31131.1 GDIQ01266766 JAJ84958.1 LRGB01001361 KZS12718.1 GDIQ01066408 JAN28329.1 GDIQ01235079 JAK16646.1 GDIQ01266465 GDIQ01144856 GDIQ01143364 JAJ85259.1 AB728502 BAM21526.1 PYGN01000508 PSN45176.1 GGMR01000593 MBY13212.1 KJ152557 AHN85840.1 ABLF02010960 GDIQ01197245 JAK54480.1 GDIP01185115 JAJ38287.1 GDIQ01051407 JAN43330.1 GDIP01054041 JAM49674.1 GDIP01156935 JAJ66467.1 GDIQ01219742 JAK31983.1 GL732667 EFX67831.1 GDIQ01169869 JAK81856.1 KZ288232 PBC31518.1 GDIQ01069507 JAN25230.1 KK116287 KFM67340.1 AB897810 BAO52003.1 GL445346 EFN89747.1 KQ414632 KOC67112.1 KQ981490 KYN41069.1 GDIQ01149313 JAL02413.1 KQ434998 KZC13267.1 KQ979851 KYN18841.1 IAAA01020797 LAA08302.1 KQ976401 KYM92530.1 ADTU01013248 KQ982696 KYQ51984.1 KQ978023 KYM97876.1 GBYB01002015 JAG71782.1 GDIQ01198644 JAK53081.1 GACK01008627 JAA56407.1 NNAY01000390 OXU28715.1 KQ435922 KOX68655.1 GL435311 EFN73526.1 GL888182 EGI65668.1 GL767671 EFZ13313.1 CAEY01001591 GECU01026383 JAS81323.1 KQ971318 KYB28963.1 BK005823 DAA05780.1 GDIP01232479 JAI90922.1
KP657630 AKL78855.1 KQ461154 KPJ08590.1 AGBW02014614 OWR41334.1 JTDY01000066 KOB79082.1 GEBQ01024543 JAT15434.1 GEDC01000504 JAS36794.1 GEDC01026310 JAS10988.1 JQ316454 AFO63077.1 JQ316456 AFO63079.1 JQ316457 AFO63080.1 JQ316453 JQ316455 AFO63076.1 AFO63078.1 NEVH01024952 PNF16319.1 KK853274 KDR09110.1 GFXV01001510 MBW13315.1 GDIQ01023602 JAN71135.1 GDIQ01097519 GDIQ01095784 GDIQ01031867 JAN62870.1 GDIQ01178047 JAK73678.1 GDIQ01236230 JAK15495.1 GDIQ01176392 JAK75333.1 GDIQ01160609 JAK91116.1 AB897808 BAO52001.1 GDIQ01162070 JAK89655.1 GDIQ01154421 JAK97304.1 GDIQ01060043 JAN34694.1 GDIQ01235080 JAK16645.1 GDIQ01127815 JAL23911.1 GDIQ01062186 JAN32551.1 GDIQ01179489 JAK72236.1 GDIQ01137464 JAL14262.1 GDIQ01141812 JAL09914.1 GDIQ01180900 GDIQ01067901 JAK70825.1 GDIQ01174142 GDIQ01057408 JAN37329.1 GDIQ01156905 JAK94820.1 GDIQ01160608 JAK91117.1 GDIQ01158906 JAK92819.1 GDIQ01131760 GDIQ01130468 JAL19966.1 GDIQ01144857 GDIQ01082337 GDIQ01042848 JAL06869.1 GDIQ01107474 JAL44252.1 GDIQ01083732 JAN11005.1 GDIQ01151526 JAL00200.1 GDIQ01030623 JAN64114.1 DS235171 EEB12867.1 GDIP01032747 JAM70968.1 GDIQ01229285 JAK22440.1 GDIQ01179490 JAK72235.1 GDIQ01180901 JAK70824.1 GDIQ01063606 JAN31131.1 GDIQ01266766 JAJ84958.1 LRGB01001361 KZS12718.1 GDIQ01066408 JAN28329.1 GDIQ01235079 JAK16646.1 GDIQ01266465 GDIQ01144856 GDIQ01143364 JAJ85259.1 AB728502 BAM21526.1 PYGN01000508 PSN45176.1 GGMR01000593 MBY13212.1 KJ152557 AHN85840.1 ABLF02010960 GDIQ01197245 JAK54480.1 GDIP01185115 JAJ38287.1 GDIQ01051407 JAN43330.1 GDIP01054041 JAM49674.1 GDIP01156935 JAJ66467.1 GDIQ01219742 JAK31983.1 GL732667 EFX67831.1 GDIQ01169869 JAK81856.1 KZ288232 PBC31518.1 GDIQ01069507 JAN25230.1 KK116287 KFM67340.1 AB897810 BAO52003.1 GL445346 EFN89747.1 KQ414632 KOC67112.1 KQ981490 KYN41069.1 GDIQ01149313 JAL02413.1 KQ434998 KZC13267.1 KQ979851 KYN18841.1 IAAA01020797 LAA08302.1 KQ976401 KYM92530.1 ADTU01013248 KQ982696 KYQ51984.1 KQ978023 KYM97876.1 GBYB01002015 JAG71782.1 GDIQ01198644 JAK53081.1 GACK01008627 JAA56407.1 NNAY01000390 OXU28715.1 KQ435922 KOX68655.1 GL435311 EFN73526.1 GL888182 EGI65668.1 GL767671 EFZ13313.1 CAEY01001591 GECU01026383 JAS81323.1 KQ971318 KYB28963.1 BK005823 DAA05780.1 GDIP01232479 JAI90922.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
UP000235965
+ More
UP000027135 UP000192223 UP000009046 UP000076858 UP000245037 UP000007819 UP000000305 UP000242457 UP000054359 UP000002358 UP000008237 UP000053825 UP000078541 UP000076502 UP000078492 UP000078540 UP000005205 UP000075809 UP000078542 UP000215335 UP000053105 UP000000311 UP000007755 UP000015104 UP000007266 UP000005203
UP000027135 UP000192223 UP000009046 UP000076858 UP000245037 UP000007819 UP000000305 UP000242457 UP000054359 UP000002358 UP000008237 UP000053825 UP000078541 UP000076502 UP000078492 UP000078540 UP000005205 UP000075809 UP000078542 UP000215335 UP000053105 UP000000311 UP000007755 UP000015104 UP000007266 UP000005203
Interpro
IPR000323
Cu2_ascorb_mOase_N
+ More
IPR020611 Cu2_ascorb_mOase_CS-1
IPR024548 Cu2_monoox_C
IPR008977 PHM/PNGase_F_dom_sf
IPR028460 Tbh/DBH
IPR036939 Cu2_ascorb_mOase_N_sf
IPR014784 Cu2_ascorb_mOase-like_C
IPR005018 DOMON_domain
IPR035979 RBD_domain_sf
IPR010280 U5_MeTrfase_fam
IPR029063 SAM-dependent_MTases
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR002059 CSP_DNA-bd
IPR012340 NA-bd_OB-fold
IPR011129 CSD
IPR014783 Cu2_ascorb_mOase_CS-2
IPR017441 Protein_kinase_ATP_BS
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR000719 Prot_kinase_dom
IPR020611 Cu2_ascorb_mOase_CS-1
IPR024548 Cu2_monoox_C
IPR008977 PHM/PNGase_F_dom_sf
IPR028460 Tbh/DBH
IPR036939 Cu2_ascorb_mOase_N_sf
IPR014784 Cu2_ascorb_mOase-like_C
IPR005018 DOMON_domain
IPR035979 RBD_domain_sf
IPR010280 U5_MeTrfase_fam
IPR029063 SAM-dependent_MTases
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR002059 CSP_DNA-bd
IPR012340 NA-bd_OB-fold
IPR011129 CSD
IPR014783 Cu2_ascorb_mOase_CS-2
IPR017441 Protein_kinase_ATP_BS
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR000719 Prot_kinase_dom
SUPFAM
Gene 3D
CDD
ProteinModelPortal
F1T2L6
A0A2A4IVW4
A0A194PQE1
A0A0G3VI00
A0A194QUD3
A0A212EIM8
+ More
A0A0L7LUG0 A0A1B6KW36 A0A1B6EFZ9 A0A1B6CBY6 I7C6J7 I7BWB2 I7D3M3 I7CTE1 A0A2J7PJ22 A0A067QTZ3 A0A2H8TGU2 A0A0P6H9G9 A0A1W4WN56 A0A0P6GQ74 A0A0P5LMW4 A0A0P5H9I3 A0A0P5LRQ0 A0A0P5MBJ7 W8VKK1 A0A0P5M143 A0A0P5P5B5 A0A0P6FVD3 A0A0P5H0D0 A0A0P5QZD0 A0A0P6EIS1 A0A0P5L6R5 A0A0P5NUR4 A0A0P5NQ72 A0A0P5LG82 A0A0P6EVL1 A0A0P5MNP2 A0A0P5MER8 A0A0P5M937 A0A0P5PMI4 A0A0P5PSW4 A0A0P5R021 A0A0N8DWZ2 A0A0P5MT95 A0A0P6HCP7 E0VHU4 A0A0P6ACI3 A0A0P5HAF7 A0A0P5MGX2 A0A0P5KR28 A0A0P6EZ15 A0A0P5EWS2 A0A164VW34 A0A0P6F255 A0A0P5IQN1 A0A0P5F328 D6WEK9 A0A2P8YLP8 A0A2S2N8E7 X2KVP5 J9K380 A0A0P5JYZ3 A0A0P5BG39 A0A0P6FB14 A0A0P5YSU2 A0A0P5DFR6 A0A0P5IC69 E9HK72 A0A0P5LGM6 A0A2A3EII7 A0A0P6EWW9 A0A087TQF1 W8VTH9 K7J3J6 E2B3J5 A0A0L7R8A1 A0A195FM60 A0A0P5MYZ9 A0A154PNF6 A0A151J6J3 A0A2L2YJK8 A0A195BW72 A0A158NEC4 A0A151WVS2 A0A195CCB6 A0A0C9R1Y8 A0A0P5JW18 L7LXT4 A0A232FDF1 A0A0M8ZPV4 E1ZZB1 F4WJ90 E9J1G9 T1K603 A0A1B6I325 A0A139WM35 Q147S1 A0A0P4YDU6
A0A0L7LUG0 A0A1B6KW36 A0A1B6EFZ9 A0A1B6CBY6 I7C6J7 I7BWB2 I7D3M3 I7CTE1 A0A2J7PJ22 A0A067QTZ3 A0A2H8TGU2 A0A0P6H9G9 A0A1W4WN56 A0A0P6GQ74 A0A0P5LMW4 A0A0P5H9I3 A0A0P5LRQ0 A0A0P5MBJ7 W8VKK1 A0A0P5M143 A0A0P5P5B5 A0A0P6FVD3 A0A0P5H0D0 A0A0P5QZD0 A0A0P6EIS1 A0A0P5L6R5 A0A0P5NUR4 A0A0P5NQ72 A0A0P5LG82 A0A0P6EVL1 A0A0P5MNP2 A0A0P5MER8 A0A0P5M937 A0A0P5PMI4 A0A0P5PSW4 A0A0P5R021 A0A0N8DWZ2 A0A0P5MT95 A0A0P6HCP7 E0VHU4 A0A0P6ACI3 A0A0P5HAF7 A0A0P5MGX2 A0A0P5KR28 A0A0P6EZ15 A0A0P5EWS2 A0A164VW34 A0A0P6F255 A0A0P5IQN1 A0A0P5F328 D6WEK9 A0A2P8YLP8 A0A2S2N8E7 X2KVP5 J9K380 A0A0P5JYZ3 A0A0P5BG39 A0A0P6FB14 A0A0P5YSU2 A0A0P5DFR6 A0A0P5IC69 E9HK72 A0A0P5LGM6 A0A2A3EII7 A0A0P6EWW9 A0A087TQF1 W8VTH9 K7J3J6 E2B3J5 A0A0L7R8A1 A0A195FM60 A0A0P5MYZ9 A0A154PNF6 A0A151J6J3 A0A2L2YJK8 A0A195BW72 A0A158NEC4 A0A151WVS2 A0A195CCB6 A0A0C9R1Y8 A0A0P5JW18 L7LXT4 A0A232FDF1 A0A0M8ZPV4 E1ZZB1 F4WJ90 E9J1G9 T1K603 A0A1B6I325 A0A139WM35 Q147S1 A0A0P4YDU6
PDB
4ZEL
E-value=2.82846e-119,
Score=1098
Ontologies
PATHWAY
GO
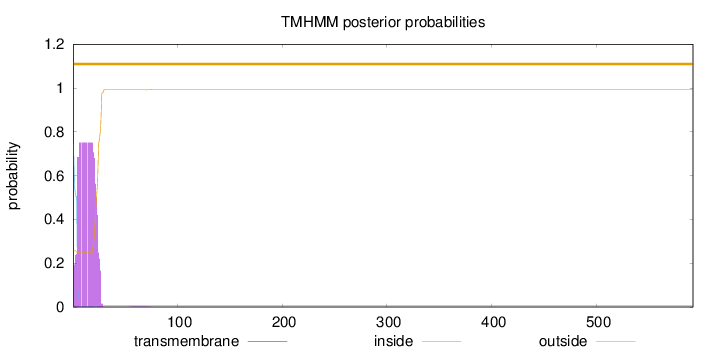
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.621979
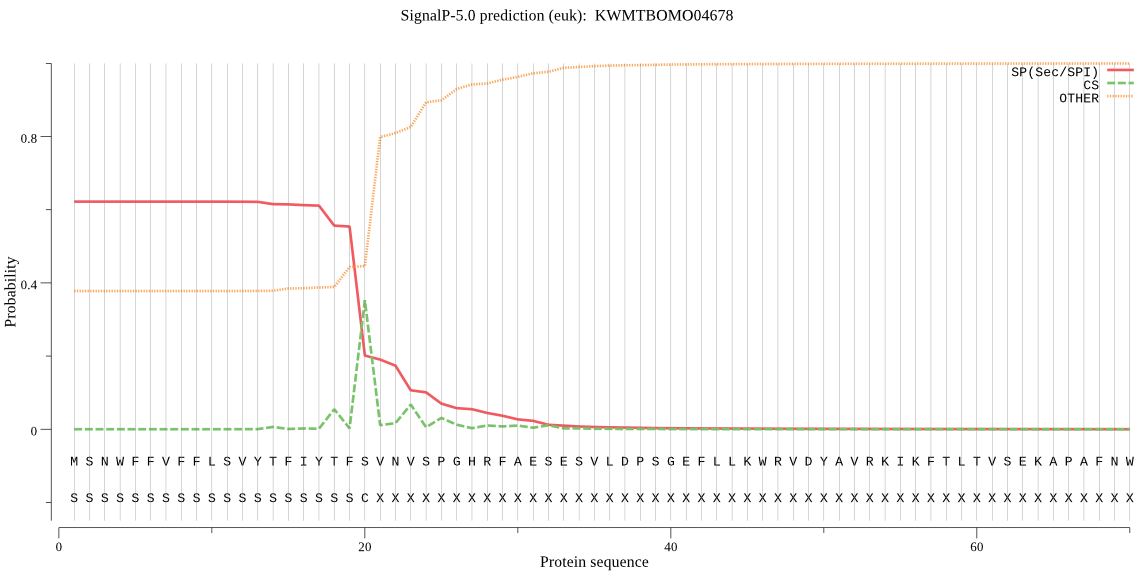
Length:
592
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
15.36485
Exp number, first 60 AAs:
15.34309
Total prob of N-in:
0.74281
POSSIBLE N-term signal
sequence
outside
1 - 592
Population Genetic Test Statistics
Pi
311.379368
Theta
195.692829
Tajima's D
2.364925
CLR
0.204443
CSRT
0.931753412329384
Interpretation
Uncertain
