Gene
KWMTBOMO04675 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005288
Annotation
PREDICTED:_FACT_complex_subunit_Ssrp1_[Amyelois_transitella]
Full name
FACT complex subunit SSRP1
Transcription factor
Location in the cell
Nuclear Reliability : 3.903
Sequence
CDS
ATGGTACCAGGGAGGTTAAAAATGACAGACCAAAACATAATTTTCAAGAATAGTAAAACCGGCAAAGTTGAACAAATATCAGCTAATGACATTGAGCTGGTGAATTTCCAGAAGTTCATTGGTTCGTGGGGCTTACGTTTGTTTCTCAAAAATGGTACTTTGCACAGATACGGAGGTTTTAAAGAAGGTGAGCAAGAGAAGGTTGCAAAATTCTTCAAAGCCAATTATAATAAAGACATGCTAGAGAAAGAGTTATCTCTAAAAGGTTGGAACTGGGGTACAGCTAAATTTAATGGAGCAGTACTAAGTTTCAATGTTGGTACAAACACCGCTTTTGAAATACCATTGCATTATGTCTCTCAATGCAATACAGGAAAGAATGAAGTCACACTCGAATTTCATCAGAATGATGACACACCTGTATCATTGATGGAAATGCGCTTCCACATTCCCACCAGTGAAGTGGCCAATGATTTGGATGCAGTTGAAGCATTCCACCAACAAGTAATGAACAAAGCGAGTGTTATTTCAGTTTCTGGTGATGCTATTGCTATATTCAGAGAACTACAATGTTTGACACCTCGTGGTCGTTATGATATCAAGGTATTCCAGACATTCTTTCAGCTTCACGGTAAAACCTTTGACTACAAGATCCCTATGTCGACAGTTCTCCGTTTATTCTTATTGCCTCACAAGGATACGAGGCAAATGTTCTTTGTCGTTTCCTTGGATCCACCGATCAAACAGGGTCAAACAAGGTACCATTATTTAGTGCTGTTGTTTGGTATTGAGGAGGAGACAACTGTAGAACTTCCTTTTACAGAGGAAGAATTGAAAGACAAATATGAAGGTAAAATAACAAAGGAATTATCTGGACCGACATACGAAGTTCTAGCCAAAATCATGAAGGTCATCATCAATAGAAGGGTAACAGGTCCTGGTGACTTTTTAGGACACCACAAGACCCCGGCCATAGCGTGTTCATACAAGGCGGCGGCAGGCTATCTGTATCCCCTAGACAAAGGTTTCATATACGTTCACAAGCCACCAGTACACATACGATTTGAGGAGATTGCGTCTGTGAACTTCGCTCGAGGGGGCGCTTCGTCGACGAAATCATTTGATTTCGAAATAGAATTGAAGTTGGGCAGCGTGCACACGTTTAGCAGTATCGAGAAGGGAGAATATGACAAACTATTCAATTATATATCTTCGAAGAAATTACATGTCAAGAATACTGGAAAGAATGACAAAGCCCTGTACGACGATGACTTCGGCGACTCGGACACGGAGAAGGAACCGGACGCGTATCTGGAGCGCGTGAAGGCGGAGGCGAAGGAGCGCGAGTCGGACGACAGCGACGGCTCCGACGAGAGCACCGACGAGGACTTCAACCCCGACAAGGCCAAGGAGAGCGACGTCGCTGAAGAATACGACACGAACGCCTCATCTTCAGAGGACTCCGACGCGTCAGGCGGCTCCGGCTCCGGTGATAGCAAGAAGGAGAAAGAAAAAGAAAAGAAGAAAGAGAAAAAACCTAAGAAAGCTAAGACTATTTCCGAAGCGCCACGTAAGCGTAAAGAGAAGTCTAAGAAACGCGAGAAGGACGTGAACGCGCCAAAGCGGCCGGCGACCGCCTTCATGCTGTGGCTCAACGAAAACAGAAAGAAAATCATTGAAGAGAACCCCGGGATCAAGGTCACAGAGGTCGCCAAGCGCGCGGGAGAACTCTGGAGGGACCTGAAAGACAAATCGGAATGGGAAGAGAAAGCCAACAAAGCGAAAGAAGAATACAATGCCGCAATGAAGAAGTATAAAGACAGCGGCGCGGCCGACGAGTTCAAACAGAAAAAGAAACAGGCTGAAAAAGAACGAAAGACAGCAGAGAAGAAGACCAAAGCTCCTCCAGCAAAGAAGACTCCTGCCGCCGTTCCCACCGGGAAGTTCACCAGCAAAGAGTTTATAGAAGACGACGATAGCTCCTCCGACTCCGACAGCAATAAGAAGAAGGAGAAGAAGGACACTAAAGACACAAAGAAAGATAAAGAAAAATCTAAGAGTTCGTCTTCCGAAGCCGAGGCATCATCGGCGTCTGGAAACGACAGCGGCAGTGATAGTAATTAA
Protein
MVPGRLKMTDQNIIFKNSKTGKVEQISANDIELVNFQKFIGSWGLRLFLKNGTLHRYGGFKEGEQEKVAKFFKANYNKDMLEKELSLKGWNWGTAKFNGAVLSFNVGTNTAFEIPLHYVSQCNTGKNEVTLEFHQNDDTPVSLMEMRFHIPTSEVANDLDAVEAFHQQVMNKASVISVSGDAIAIFRELQCLTPRGRYDIKVFQTFFQLHGKTFDYKIPMSTVLRLFLLPHKDTRQMFFVVSLDPPIKQGQTRYHYLVLLFGIEEETTVELPFTEEELKDKYEGKITKELSGPTYEVLAKIMKVIINRRVTGPGDFLGHHKTPAIACSYKAAAGYLYPLDKGFIYVHKPPVHIRFEEIASVNFARGGASSTKSFDFEIELKLGSVHTFSSIEKGEYDKLFNYISSKKLHVKNTGKNDKALYDDDFGDSDTEKEPDAYLERVKAEAKERESDDSDGSDESTDEDFNPDKAKESDVAEEYDTNASSSEDSDASGGSGSGDSKKEKEKEKKKEKKPKKAKTISEAPRKRKEKSKKREKDVNAPKRPATAFMLWLNENRKKIIEENPGIKVTEVAKRAGELWRDLKDKSEWEEKANKAKEEYNAAMKKYKDSGAADEFKQKKKQAEKERKTAEKKTKAPPAKKTPAAVPTGKFTSKEFIEDDDSSSDSDSNKKKEKKDTKDTKKDKEKSKSSSSEAEASSASGNDSGSDSN
Summary
Description
Component of the FACT complex, a general chromatin factor that acts to reorganize nucleosomes. The FACT complex is involved in multiple processes that require DNA as a template such as mRNA elongation, DNA replication and DNA repair. During transcription elongation the FACT complex acts as a histone chaperone that both destabilizes and restores nucleosomal structure. It facilitates the passage of RNA polymerase II and transcription by promoting the dissociation of one histone H2A-H2B dimer from the nucleosome, then subsequently promotes the reestablishment of the nucleosome following the passage of RNA polymerase II.
Similarity
Belongs to the SSRP1 family.
Uniprot
H9J6Z6
A0A2H1VJZ7
A0A2A4J926
A0A194QSY9
A0A194PQS3
A0A1E1W699
+ More
A0A212ENB7 S4PXG7 E2AEI3 F4WJ99 A0A0J7L690 A0A067R232 A0A026W7R0 A0A158NEB6 D6X1B7 A0A0T6B816 A0A088ASB6 A0A2Z5U5X6 A0A2P8XHV6 A0A1Y1L9T7 E2BLQ9 A0A2A3ERW1 A0A310SQT6 A0A1L8DGV0 A0A232F9X2 K7J3D9 A0A0C9R8P1 A0A1B6EEE8 N6US47 A0A1W4X9F1 A0A1B6I9E0 A0A1B6FK22 A0A1B6G5T8 A0A0A9X2W4 Q0IEB2 A0A034WBT1 W8BD77 A0A0V0G502 A0A224X7K1 A0A0K8UFD2 A0A2S2P9X7 A0A1I8PFQ6 A0A2H8TUF8 A0A069DZJ5 R4FNX0 A0A151WSH0 A0A1B0G7P8 B3NPS4 B4PA67 A0A1I8MR81 T1PQ22 E0VDU0 A0A1S3DIE7 A0A1D2MYP6 A0A0P5IFZ6
A0A212ENB7 S4PXG7 E2AEI3 F4WJ99 A0A0J7L690 A0A067R232 A0A026W7R0 A0A158NEB6 D6X1B7 A0A0T6B816 A0A088ASB6 A0A2Z5U5X6 A0A2P8XHV6 A0A1Y1L9T7 E2BLQ9 A0A2A3ERW1 A0A310SQT6 A0A1L8DGV0 A0A232F9X2 K7J3D9 A0A0C9R8P1 A0A1B6EEE8 N6US47 A0A1W4X9F1 A0A1B6I9E0 A0A1B6FK22 A0A1B6G5T8 A0A0A9X2W4 Q0IEB2 A0A034WBT1 W8BD77 A0A0V0G502 A0A224X7K1 A0A0K8UFD2 A0A2S2P9X7 A0A1I8PFQ6 A0A2H8TUF8 A0A069DZJ5 R4FNX0 A0A151WSH0 A0A1B0G7P8 B3NPS4 B4PA67 A0A1I8MR81 T1PQ22 E0VDU0 A0A1S3DIE7 A0A1D2MYP6 A0A0P5IFZ6
Pubmed
EMBL
BABH01014064
ODYU01002808
SOQ40762.1
NWSH01002536
PCG68034.1
KQ461154
+ More
KPJ08587.1 KQ459595 KPI95662.1 GDQN01008585 JAT82469.1 AGBW02013681 OWR42984.1 GAIX01005068 JAA87492.1 GL438862 EFN68140.1 GL888182 EGI65677.1 LBMM01000514 KMQ98166.1 KK853076 KDR11732.1 KK107356 QOIP01000010 EZA52038.1 RLU17467.1 ADTU01013241 ADTU01013242 KQ971372 EFA10605.2 LJIG01009371 KRT83213.1 FX985766 BBA93653.1 PYGN01002085 PSN31544.1 GEZM01066473 JAV67857.1 GL449036 EFN83393.1 KZ288191 PBC34430.1 KQ761155 OAD58135.1 GFDF01008489 JAV05595.1 NNAY01000645 OXU27223.1 GBYB01004505 GBYB01004508 GBYB01011733 JAG74272.1 JAG74275.1 JAG81500.1 GEDC01001005 JAS36293.1 APGK01018910 KB740085 ENN81542.1 GECU01024174 JAS83532.1 GECZ01019239 JAS50530.1 GECZ01011971 JAS57798.1 GBHO01029265 GBRD01005968 JAG14339.1 JAG59853.1 CH477764 EAT36499.1 GAKP01007190 JAC51762.1 GAMC01009923 JAB96632.1 GECL01003009 JAP03115.1 GFTR01008029 JAW08397.1 GDHF01026932 JAI25382.1 GGMR01013616 MBY26235.1 GFXV01006108 MBW17913.1 GBGD01000530 JAC88359.1 ACPB03000491 GAHY01000398 JAA77112.1 KQ982769 KYQ50820.1 CCAG010016890 CH954179 EDV56865.1 CM000158 EDW92393.1 KA650208 AFP64837.1 DS235088 EEB11546.1 LJIJ01000385 ODM98071.1 GDIQ01215237 JAK36488.1
KPJ08587.1 KQ459595 KPI95662.1 GDQN01008585 JAT82469.1 AGBW02013681 OWR42984.1 GAIX01005068 JAA87492.1 GL438862 EFN68140.1 GL888182 EGI65677.1 LBMM01000514 KMQ98166.1 KK853076 KDR11732.1 KK107356 QOIP01000010 EZA52038.1 RLU17467.1 ADTU01013241 ADTU01013242 KQ971372 EFA10605.2 LJIG01009371 KRT83213.1 FX985766 BBA93653.1 PYGN01002085 PSN31544.1 GEZM01066473 JAV67857.1 GL449036 EFN83393.1 KZ288191 PBC34430.1 KQ761155 OAD58135.1 GFDF01008489 JAV05595.1 NNAY01000645 OXU27223.1 GBYB01004505 GBYB01004508 GBYB01011733 JAG74272.1 JAG74275.1 JAG81500.1 GEDC01001005 JAS36293.1 APGK01018910 KB740085 ENN81542.1 GECU01024174 JAS83532.1 GECZ01019239 JAS50530.1 GECZ01011971 JAS57798.1 GBHO01029265 GBRD01005968 JAG14339.1 JAG59853.1 CH477764 EAT36499.1 GAKP01007190 JAC51762.1 GAMC01009923 JAB96632.1 GECL01003009 JAP03115.1 GFTR01008029 JAW08397.1 GDHF01026932 JAI25382.1 GGMR01013616 MBY26235.1 GFXV01006108 MBW17913.1 GBGD01000530 JAC88359.1 ACPB03000491 GAHY01000398 JAA77112.1 KQ982769 KYQ50820.1 CCAG010016890 CH954179 EDV56865.1 CM000158 EDW92393.1 KA650208 AFP64837.1 DS235088 EEB11546.1 LJIJ01000385 ODM98071.1 GDIQ01215237 JAK36488.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000000311
+ More
UP000007755 UP000036403 UP000027135 UP000053097 UP000279307 UP000005205 UP000007266 UP000005203 UP000245037 UP000008237 UP000242457 UP000215335 UP000002358 UP000019118 UP000192223 UP000008820 UP000095300 UP000015103 UP000075809 UP000092444 UP000008711 UP000002282 UP000095301 UP000009046 UP000079169 UP000094527
UP000007755 UP000036403 UP000027135 UP000053097 UP000279307 UP000005205 UP000007266 UP000005203 UP000245037 UP000008237 UP000242457 UP000215335 UP000002358 UP000019118 UP000192223 UP000008820 UP000095300 UP000015103 UP000075809 UP000092444 UP000008711 UP000002282 UP000095301 UP000009046 UP000079169 UP000094527
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9J6Z6
A0A2H1VJZ7
A0A2A4J926
A0A194QSY9
A0A194PQS3
A0A1E1W699
+ More
A0A212ENB7 S4PXG7 E2AEI3 F4WJ99 A0A0J7L690 A0A067R232 A0A026W7R0 A0A158NEB6 D6X1B7 A0A0T6B816 A0A088ASB6 A0A2Z5U5X6 A0A2P8XHV6 A0A1Y1L9T7 E2BLQ9 A0A2A3ERW1 A0A310SQT6 A0A1L8DGV0 A0A232F9X2 K7J3D9 A0A0C9R8P1 A0A1B6EEE8 N6US47 A0A1W4X9F1 A0A1B6I9E0 A0A1B6FK22 A0A1B6G5T8 A0A0A9X2W4 Q0IEB2 A0A034WBT1 W8BD77 A0A0V0G502 A0A224X7K1 A0A0K8UFD2 A0A2S2P9X7 A0A1I8PFQ6 A0A2H8TUF8 A0A069DZJ5 R4FNX0 A0A151WSH0 A0A1B0G7P8 B3NPS4 B4PA67 A0A1I8MR81 T1PQ22 E0VDU0 A0A1S3DIE7 A0A1D2MYP6 A0A0P5IFZ6
A0A212ENB7 S4PXG7 E2AEI3 F4WJ99 A0A0J7L690 A0A067R232 A0A026W7R0 A0A158NEB6 D6X1B7 A0A0T6B816 A0A088ASB6 A0A2Z5U5X6 A0A2P8XHV6 A0A1Y1L9T7 E2BLQ9 A0A2A3ERW1 A0A310SQT6 A0A1L8DGV0 A0A232F9X2 K7J3D9 A0A0C9R8P1 A0A1B6EEE8 N6US47 A0A1W4X9F1 A0A1B6I9E0 A0A1B6FK22 A0A1B6G5T8 A0A0A9X2W4 Q0IEB2 A0A034WBT1 W8BD77 A0A0V0G502 A0A224X7K1 A0A0K8UFD2 A0A2S2P9X7 A0A1I8PFQ6 A0A2H8TUF8 A0A069DZJ5 R4FNX0 A0A151WSH0 A0A1B0G7P8 B3NPS4 B4PA67 A0A1I8MR81 T1PQ22 E0VDU0 A0A1S3DIE7 A0A1D2MYP6 A0A0P5IFZ6
PDB
5UMS
E-value=1.30676e-99,
Score=930
Ontologies
KEGG
GO
GO:0005694
GO:0006281
GO:0005634
GO:0006260
GO:0003677
GO:0035101
GO:0031491
GO:0000790
GO:0042393
GO:0030838
GO:0005705
GO:0046982
GO:0030707
GO:0043621
GO:0005703
GO:0001672
GO:0031492
GO:0001933
GO:0005730
GO:0070087
GO:0005829
GO:0051101
GO:0030131
GO:0005622
GO:0009058
GO:0005089
GO:0035023
GO:0005515
GO:0016742
GO:0003824
Topology
Subcellular location
Nucleus
Chromosome
Chromosome
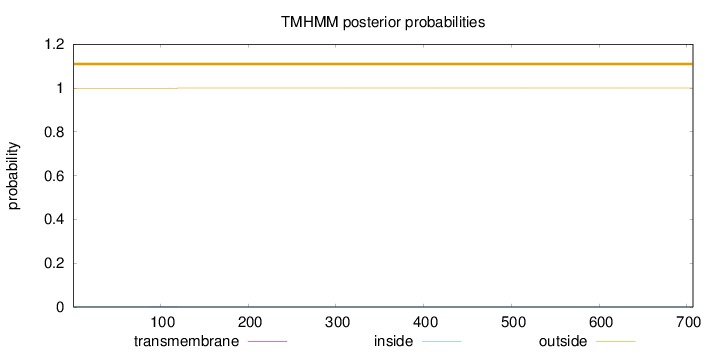
Length:
707
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0119
Exp number, first 60 AAs:
0.00084
Total prob of N-in:
0.00073
outside
1 - 707
Population Genetic Test Statistics
Pi
188.993158
Theta
180.359091
Tajima's D
0.138936
CLR
0.514907
CSRT
0.404029798510074
Interpretation
Uncertain
