Gene
KWMTBOMO04672
Pre Gene Modal
BGIBMGA005520
Annotation
PREDICTED:_ADP-ribosylation_factor-binding_protein_GGA1_isoform_X2_[Bombyx_mori]
Location in the cell
Golgi Reliability : 1.396 Nuclear Reliability : 1.97
Sequence
CDS
ATGAATATTACACAAACAAGCTTAGAAGCACTTATTTTAAAAGCTACTCACGAATCATTACCGGCTCCCGATGCAGCGGCTGTAGAAGCATTCTGCACGATTATGCGAGACTCTGCCGAAGGTCCCCAACTGGCAGCGCAAGCTCTGGCCACACGTATCCACAGTCCGAAGGCCCGTGAAGCGCTACTTGCATTGATGATGTTAGATCGCTGTATGCGGCGATGCGACACGCCTTTCCATGCGGAAATCGGAAAATTTCGCTTTCTTAATGAAATGATAAAATTAGTGTCACCAAAATATTATGCAGATCGTACAGCGCCAGAAGTAAAGACAAGGGTTTTGCAGTTGTTCCATGCATGGTCTATAGAATATCATAAGGAGTTTAAATTCAAAGTAGCATATGAGATGTTAAAAAATCAAGGTGTGATAAGAGAGACACCTCCTCCTTTACCGCCAGATGATACACCTCCAGTACATAATCGAGCAAAAAGTACTATATTTGAAGATGAAGAGAAATCTAAACTGTTACAAAAATTATTACAAAGTAAAAAATCAGAAGATTTACAACATGCCAATAGACTTATAAAAACTATGGTTAGAGAGGTAGAACGTCAGAATGAGGCAAACAGCAGACGCGCACAAGAAGTTAGTGCGGCTCTAGATAGTGCCGGGCTATTACGTGACATGCTACTGGCAGCCAGTGATGCTTCATCGGACGAGGAGCAACTGATCCACGAGCTACACTCAACTTGCATTAGACTTAGGCCCATTCTGCAGCAACTGGCTGACAATGACACACAAGCAGAATATCTTAGTGATATTTTACATGCCAGTGATGTGATAGATGAAGTCTGCGAGAGGTACAATGTTTTCTTAATTCAGAAAAAAGCAAATGAACAAATGACAGAACAGGCATCGTTGAGCGGTGAAAGCTTACTTGATTTTGCTGGAGCAAGTACTACTTACATTCGAAAGAATGTGGCCCCGAAGAAAGATGAAAAGCCAGCAAATGTTATTGATGAACTAGGCGATATATTCGCTGCTAGCAGTGAAAGCACTAATAATATTGCTGAACCATTAAAGCCTGTTGCATTGATGCCCACTGCAGATGATGTCGACTTGTTGGGAGACAGATCTAATAAACATGAAGGTTGGAACGAATTGGATTCCCTTGGAGAAGAACTATTAAAACAATCTCTACCAGAGAATGCTAAACGGATTGAAAACTTTAATAGTAAAACATCAACAAAGGTACCAATGAACGCATTGGCCTCACCAAAACATGATGTGAATTTACAGAACAACGGCGCATTACTAGATTTAGATTTCTTCACCAAGAAACCTCAGGAATGTAAACCTGTAACTGCGTCAACTTCTAAAGATGTGACACCCAGCGATGACATCATGGTGGATATTAGCACAGACGTTACTGATGCAACTCTCGTTTTGAACAACAATACTGAGTGTAATTTAAATAATATATTGGATTTCGGATTAGACAATGCCAAAAGTGAAACAGTAATTGATAAGGAAACTAGTGTTATTGAAAAGAAAGAAGTAAATCATAAGAATAACGAAGTGAAACCGTTGACTGATATAAATGTTACGTTGCAAAACGTAAGACCCAGTAAATTACAGCCGCTTACAGTGTTTGAAGAGGAAGAAGGTGTGACAGTTGTTTTACATTTCTGCCAAGATAAACCGAGGCAAGATGTTAATGTTTTAGTCATATCGACTACAAGTAAGAATAAATTGCCCATCGAAGATTATAAATTCCAAAGTGTAGTTCCTAAAGGTTGTAAAGTTCGGCTACTGACACCGTCGGGGACGTCACTGCCGGCGTACAATCCGTTCCTGCCGCCGCCAGCCCTGACGCAGGTCATGCTGCTCGCGAGGCCGCCTTCTATGAATAACATCAGCCTGAAATTCGTTATCACGTACACGTGCGACGAGGAGGTGTATTCAGAAATTGGCGAAGTCAAAGAACTGCCTCTAGAGAAAATCTAA
Protein
MNITQTSLEALILKATHESLPAPDAAAVEAFCTIMRDSAEGPQLAAQALATRIHSPKAREALLALMMLDRCMRRCDTPFHAEIGKFRFLNEMIKLVSPKYYADRTAPEVKTRVLQLFHAWSIEYHKEFKFKVAYEMLKNQGVIRETPPPLPPDDTPPVHNRAKSTIFEDEEKSKLLQKLLQSKKSEDLQHANRLIKTMVREVERQNEANSRRAQEVSAALDSAGLLRDMLLAASDASSDEEQLIHELHSTCIRLRPILQQLADNDTQAEYLSDILHASDVIDEVCERYNVFLIQKKANEQMTEQASLSGESLLDFAGASTTYIRKNVAPKKDEKPANVIDELGDIFAASSESTNNIAEPLKPVALMPTADDVDLLGDRSNKHEGWNELDSLGEELLKQSLPENAKRIENFNSKTSTKVPMNALASPKHDVNLQNNGALLDLDFFTKKPQECKPVTASTSKDVTPSDDIMVDISTDVTDATLVLNNNTECNLNNILDFGLDNAKSETVIDKETSVIEKKEVNHKNNEVKPLTDINVTLQNVRPSKLQPLTVFEEEEGVTVVLHFCQDKPRQDVNVLVISTTSKNKLPIEDYKFQSVVPKGCKVRLLTPSGTSLPAYNPFLPPPALTQVMLLARPPSMNNISLKFVITYTCDEEVYSEIGEVKELPLEKI
Summary
Uniprot
H9J7M8
A0A212ENF3
A0A2A4J111
A0A2H1VL39
A0A194PSC6
A0A2A4IYV8
+ More
A0A2A4IZB5 A0A194QTE7 A0A1Y1L6Y5 A0A1Y1LBZ0 D6X0M6 A0A154P5A2 A0A0N0BJA1 E2BSS0 A0A224XL36 E0VCT8 U4U3C3 N6TMG5 A0A0T6BHI0 A0A146LR51 U4UEP3 N6TQ37 A0A1L8GNW8 A0A3Q2E3E2 A0A147A1Q6 A0A3Q3LGL8 Q28E38 A0A3Q3RX55 A0A3Q3LEB0 A0A3B5AQU4 A0A3Q1G8A3 A0A146MX01 A0A1A7YL76 A0A1A7WE35 A0A1A8M111 A0A1A8QXV8 A0A1A8LE52 A0A3Q0RL69 A0A1A8KDP0 A0A1A8GV29 A0A1A8FFW3 A0A1A8ADZ3 A0A2I4CYZ6 A0A2U3ZR41 A0A2U3YBB0 A0A3S2P8F2 A0A3Q1JLN3 A0A3B5BAQ4 A0A3Q1HUM1
A0A2A4IZB5 A0A194QTE7 A0A1Y1L6Y5 A0A1Y1LBZ0 D6X0M6 A0A154P5A2 A0A0N0BJA1 E2BSS0 A0A224XL36 E0VCT8 U4U3C3 N6TMG5 A0A0T6BHI0 A0A146LR51 U4UEP3 N6TQ37 A0A1L8GNW8 A0A3Q2E3E2 A0A147A1Q6 A0A3Q3LGL8 Q28E38 A0A3Q3RX55 A0A3Q3LEB0 A0A3B5AQU4 A0A3Q1G8A3 A0A146MX01 A0A1A7YL76 A0A1A7WE35 A0A1A8M111 A0A1A8QXV8 A0A1A8LE52 A0A3Q0RL69 A0A1A8KDP0 A0A1A8GV29 A0A1A8FFW3 A0A1A8ADZ3 A0A2I4CYZ6 A0A2U3ZR41 A0A2U3YBB0 A0A3S2P8F2 A0A3Q1JLN3 A0A3B5BAQ4 A0A3Q1HUM1
Pubmed
EMBL
BABH01014061
BABH01014062
AGBW02013681
OWR42981.1
NWSH01003929
PCG65671.1
+ More
ODYU01003149 SOQ41535.1 KQ459595 KPI95659.1 NWSH01005084 PCG64454.1 PCG64453.1 KQ461154 KPJ08584.1 GEZM01062874 JAV69443.1 GEZM01062873 JAV69445.1 KQ971371 EFA10119.2 KQ434809 KZC06358.1 KQ435719 KOX78812.1 GL450241 EFN81316.1 GFTR01007687 JAW08739.1 DS235065 EEB11194.1 KB632056 ERL88369.1 APGK01059126 APGK01059127 APGK01059128 KB741292 ENN70430.1 LJIG01000238 KRT86702.1 GDHC01009462 JAQ09167.1 ERL88370.1 ENN70431.1 CM004472 OCT85547.1 GCES01060167 GCES01014241 JAR72082.1 CR848466 CAJ82212.1 GCES01162139 JAQ24183.1 HADX01008443 SBP30675.1 HADW01002394 SBP03794.1 HAEF01010478 SBR50458.1 HAEG01014725 SBR98352.1 HAEF01005495 SBR42877.1 HAEE01010368 SBR30418.1 HAEC01007608 SBQ75746.1 HAEB01011768 SBQ58295.1 HADY01013960 SBP52445.1 CM012444 RVE69152.1
ODYU01003149 SOQ41535.1 KQ459595 KPI95659.1 NWSH01005084 PCG64454.1 PCG64453.1 KQ461154 KPJ08584.1 GEZM01062874 JAV69443.1 GEZM01062873 JAV69445.1 KQ971371 EFA10119.2 KQ434809 KZC06358.1 KQ435719 KOX78812.1 GL450241 EFN81316.1 GFTR01007687 JAW08739.1 DS235065 EEB11194.1 KB632056 ERL88369.1 APGK01059126 APGK01059127 APGK01059128 KB741292 ENN70430.1 LJIG01000238 KRT86702.1 GDHC01009462 JAQ09167.1 ERL88370.1 ENN70431.1 CM004472 OCT85547.1 GCES01060167 GCES01014241 JAR72082.1 CR848466 CAJ82212.1 GCES01162139 JAQ24183.1 HADX01008443 SBP30675.1 HADW01002394 SBP03794.1 HAEF01010478 SBR50458.1 HAEG01014725 SBR98352.1 HAEF01005495 SBR42877.1 HAEE01010368 SBR30418.1 HAEC01007608 SBQ75746.1 HAEB01011768 SBQ58295.1 HADY01013960 SBP52445.1 CM012444 RVE69152.1
Proteomes
PRIDE
Pfam
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J7M8
A0A212ENF3
A0A2A4J111
A0A2H1VL39
A0A194PSC6
A0A2A4IYV8
+ More
A0A2A4IZB5 A0A194QTE7 A0A1Y1L6Y5 A0A1Y1LBZ0 D6X0M6 A0A154P5A2 A0A0N0BJA1 E2BSS0 A0A224XL36 E0VCT8 U4U3C3 N6TMG5 A0A0T6BHI0 A0A146LR51 U4UEP3 N6TQ37 A0A1L8GNW8 A0A3Q2E3E2 A0A147A1Q6 A0A3Q3LGL8 Q28E38 A0A3Q3RX55 A0A3Q3LEB0 A0A3B5AQU4 A0A3Q1G8A3 A0A146MX01 A0A1A7YL76 A0A1A7WE35 A0A1A8M111 A0A1A8QXV8 A0A1A8LE52 A0A3Q0RL69 A0A1A8KDP0 A0A1A8GV29 A0A1A8FFW3 A0A1A8ADZ3 A0A2I4CYZ6 A0A2U3ZR41 A0A2U3YBB0 A0A3S2P8F2 A0A3Q1JLN3 A0A3B5BAQ4 A0A3Q1HUM1
A0A2A4IZB5 A0A194QTE7 A0A1Y1L6Y5 A0A1Y1LBZ0 D6X0M6 A0A154P5A2 A0A0N0BJA1 E2BSS0 A0A224XL36 E0VCT8 U4U3C3 N6TMG5 A0A0T6BHI0 A0A146LR51 U4UEP3 N6TQ37 A0A1L8GNW8 A0A3Q2E3E2 A0A147A1Q6 A0A3Q3LGL8 Q28E38 A0A3Q3RX55 A0A3Q3LEB0 A0A3B5AQU4 A0A3Q1G8A3 A0A146MX01 A0A1A7YL76 A0A1A7WE35 A0A1A8M111 A0A1A8QXV8 A0A1A8LE52 A0A3Q0RL69 A0A1A8KDP0 A0A1A8GV29 A0A1A8FFW3 A0A1A8ADZ3 A0A2I4CYZ6 A0A2U3ZR41 A0A2U3YBB0 A0A3S2P8F2 A0A3Q1JLN3 A0A3B5BAQ4 A0A3Q1HUM1
PDB
1LF8
E-value=7.42583e-30,
Score=328
Ontologies
GO
Topology
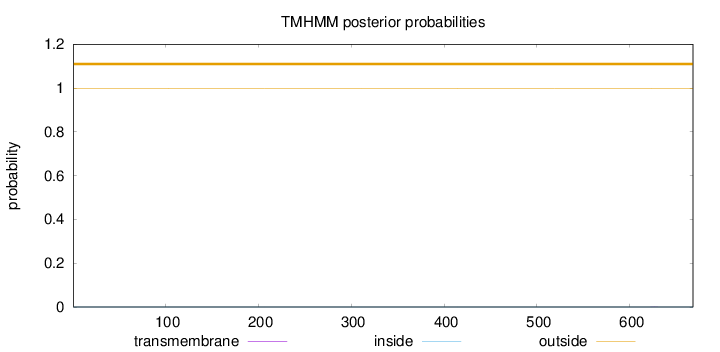
Length:
668
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00258
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00040
outside
1 - 668
Population Genetic Test Statistics
Pi
202.710998
Theta
189.469863
Tajima's D
-0.483378
CLR
0.285478
CSRT
0.244287785610719
Interpretation
Possibly Positive selection
