Gene
KWMTBOMO04671
Pre Gene Modal
BGIBMGA005291
Annotation
PREDICTED:_protein_EMRE_homolog?_mitochondrial_[Papilio_machaon]
Full name
Essential MCU regulator, mitochondrial
Location in the cell
Nuclear Reliability : 1.1
Sequence
CDS
ATGGCGATTTTTAGACTAACTCGTCTTGTGGCACAGTCCAAGACTTTAACGAAACACGAACCGATTATACATCTAAGAACTGCAACTACTACTCCAACTGGTGCTATTTTACCCGAACCACAAAAACAACCTTTTGGTTTGCTGTGTGTTGTTTTTGCTGTCGTACCGGGCTTACTAATCGGAGCTGCTATTAGTAAAAACATAGCAAATTTCCTTGAAGAAAATGAACTCTTTGTACCATCGGACGATGACGATGATGACGACTAA
Protein
MAIFRLTRLVAQSKTLTKHEPIIHLRTATTTPTGAILPEPQKQPFGLLCVVFAVVPGLLIGAAISKNIANFLEENELFVPSDDDDDDD
Summary
Description
Essential regulatory subunit of the mitochondrial calcium uniporter MCU channel, a protein that mediates calcium uptake into mitochondria.
Essential regulatory subunit of the mitochondrial calcium uniporter (mcu) channel, a protein that mediates calcium uptake into mitochondria.
Essential regulatory subunit of the mitochondrial calcium uniporter (mcu) channel, a protein that mediates calcium uptake into mitochondria.
Similarity
Belongs to the SMDT1/EMRE family.
Keywords
Calcium
Calcium transport
Complete proteome
Ion transport
Membrane
Mitochondrion
Mitochondrion inner membrane
Reference proteome
Transit peptide
Transmembrane
Transmembrane helix
Transport
Feature
chain Essential MCU regulator, mitochondrial
Uniprot
H9J6Z9
A0A2H1VMQ1
I4DJI6
A0A194QSU0
A0A212ENB3
A0A0L7L8W3
+ More
A0A2J7QXL0 E2BLJ5 A0A195BMU3 A0A195F2G1 V5GDE8 A0A151J2T5 F4WIC0 E9ISY1 A0A195CM30 A0A151XBK9 A0A026WQC6 D6X268 A0A0L7QXS6 B4P4R3 A0A310SDD0 A0A1B2AJ28 B4HNC6 B4QBV9 Q7JX57 B3NLP8 Q17ED3 A0A0N0BGU2 N6T3F5 A0A1W4UZN6 A0A1Q3FJU5 A0A1Q3FIV0 A0A182QVS4 A0A182JQZ0 B3MH49 A0A2M3ZC54 B0W2N0 T1E7K1 B4GBM0 Q290M9 A0A2A3E8X3 A0A084VD24 A0A2M3ZCK6 W5JRN2 A0A2P6KEH4 A0A0K8TPV1 B4LKW9 A0A182RL01 A0A336MPI5 A0A1D2MJW3 A0A2M4C4T6 B4J5I1 A0A1I8MES3 H2MII7 A0A3P9IBG9 A0A3P9LV58
A0A2J7QXL0 E2BLJ5 A0A195BMU3 A0A195F2G1 V5GDE8 A0A151J2T5 F4WIC0 E9ISY1 A0A195CM30 A0A151XBK9 A0A026WQC6 D6X268 A0A0L7QXS6 B4P4R3 A0A310SDD0 A0A1B2AJ28 B4HNC6 B4QBV9 Q7JX57 B3NLP8 Q17ED3 A0A0N0BGU2 N6T3F5 A0A1W4UZN6 A0A1Q3FJU5 A0A1Q3FIV0 A0A182QVS4 A0A182JQZ0 B3MH49 A0A2M3ZC54 B0W2N0 T1E7K1 B4GBM0 Q290M9 A0A2A3E8X3 A0A084VD24 A0A2M3ZCK6 W5JRN2 A0A2P6KEH4 A0A0K8TPV1 B4LKW9 A0A182RL01 A0A336MPI5 A0A1D2MJW3 A0A2M4C4T6 B4J5I1 A0A1I8MES3 H2MII7 A0A3P9IBG9 A0A3P9LV58
Pubmed
EMBL
BABH01014061
ODYU01003149
SOQ41534.1
AK401454
KQ459595
BAM18076.1
+ More
KPI95657.1 KQ461154 KPJ08583.1 AGBW02013681 OWR42980.1 JTDY01002167 KOB71968.1 NEVH01009378 PNF33326.1 GL449033 EFN83432.1 KQ976440 KYM86487.1 KQ981856 KYN34648.1 GALX01006442 JAB62024.1 KQ980404 KYN16192.1 GL888175 EGI66051.1 GL765434 EFZ16365.1 KQ977580 KYN01763.1 KQ982330 KYQ57678.1 KK107139 EZA57896.1 KQ971371 EFA09907.1 KQ414704 KOC63356.1 CM000158 EDW91686.1 KQ761039 OAD58386.1 KX531345 ANY27155.1 CH480816 EDW48408.1 CM000362 CM002911 EDX07619.1 KMY94769.1 AE013599 AY095085 CH954179 EDV55023.1 CH477283 EAT44843.1 KQ435767 KOX75281.1 APGK01054008 KB741242 KB631761 ENN72088.1 ERL85910.1 GFDL01007273 JAV27772.1 GFDL01007597 JAV27448.1 AXCN02002653 CH902619 EDV35808.1 GGFM01005386 MBW26137.1 DS231827 EDS29338.1 GAMD01002967 JAA98623.1 CH479181 EDW32278.1 CM000071 EAL25333.1 KZ288322 PBC28233.1 ATLV01011135 KE524643 KFB35868.1 GGFM01005478 MBW26229.1 ADMH02000297 ETN67067.1 MWRG01014206 PRD24725.1 GDAI01001447 JAI16156.1 CH940648 EDW60773.1 UFQT01001564 SSX31029.1 LJIJ01001044 ODM93221.1 GGFJ01011010 MBW60151.1 CH916367 EDW00744.1
KPI95657.1 KQ461154 KPJ08583.1 AGBW02013681 OWR42980.1 JTDY01002167 KOB71968.1 NEVH01009378 PNF33326.1 GL449033 EFN83432.1 KQ976440 KYM86487.1 KQ981856 KYN34648.1 GALX01006442 JAB62024.1 KQ980404 KYN16192.1 GL888175 EGI66051.1 GL765434 EFZ16365.1 KQ977580 KYN01763.1 KQ982330 KYQ57678.1 KK107139 EZA57896.1 KQ971371 EFA09907.1 KQ414704 KOC63356.1 CM000158 EDW91686.1 KQ761039 OAD58386.1 KX531345 ANY27155.1 CH480816 EDW48408.1 CM000362 CM002911 EDX07619.1 KMY94769.1 AE013599 AY095085 CH954179 EDV55023.1 CH477283 EAT44843.1 KQ435767 KOX75281.1 APGK01054008 KB741242 KB631761 ENN72088.1 ERL85910.1 GFDL01007273 JAV27772.1 GFDL01007597 JAV27448.1 AXCN02002653 CH902619 EDV35808.1 GGFM01005386 MBW26137.1 DS231827 EDS29338.1 GAMD01002967 JAA98623.1 CH479181 EDW32278.1 CM000071 EAL25333.1 KZ288322 PBC28233.1 ATLV01011135 KE524643 KFB35868.1 GGFM01005478 MBW26229.1 ADMH02000297 ETN67067.1 MWRG01014206 PRD24725.1 GDAI01001447 JAI16156.1 CH940648 EDW60773.1 UFQT01001564 SSX31029.1 LJIJ01001044 ODM93221.1 GGFJ01011010 MBW60151.1 CH916367 EDW00744.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000037510
UP000235965
+ More
UP000008237 UP000078540 UP000078541 UP000078492 UP000007755 UP000078542 UP000075809 UP000053097 UP000007266 UP000053825 UP000002282 UP000001292 UP000000304 UP000000803 UP000008711 UP000008820 UP000053105 UP000019118 UP000030742 UP000192221 UP000075886 UP000075881 UP000007801 UP000002320 UP000008744 UP000001819 UP000242457 UP000030765 UP000000673 UP000008792 UP000075900 UP000094527 UP000001070 UP000095301 UP000001038 UP000265200 UP000265180
UP000008237 UP000078540 UP000078541 UP000078492 UP000007755 UP000078542 UP000075809 UP000053097 UP000007266 UP000053825 UP000002282 UP000001292 UP000000304 UP000000803 UP000008711 UP000008820 UP000053105 UP000019118 UP000030742 UP000192221 UP000075886 UP000075881 UP000007801 UP000002320 UP000008744 UP000001819 UP000242457 UP000030765 UP000000673 UP000008792 UP000075900 UP000094527 UP000001070 UP000095301 UP000001038 UP000265200 UP000265180
Pfam
PF10161 DDDD
Interpro
IPR018782
MCU_reg
ProteinModelPortal
H9J6Z9
A0A2H1VMQ1
I4DJI6
A0A194QSU0
A0A212ENB3
A0A0L7L8W3
+ More
A0A2J7QXL0 E2BLJ5 A0A195BMU3 A0A195F2G1 V5GDE8 A0A151J2T5 F4WIC0 E9ISY1 A0A195CM30 A0A151XBK9 A0A026WQC6 D6X268 A0A0L7QXS6 B4P4R3 A0A310SDD0 A0A1B2AJ28 B4HNC6 B4QBV9 Q7JX57 B3NLP8 Q17ED3 A0A0N0BGU2 N6T3F5 A0A1W4UZN6 A0A1Q3FJU5 A0A1Q3FIV0 A0A182QVS4 A0A182JQZ0 B3MH49 A0A2M3ZC54 B0W2N0 T1E7K1 B4GBM0 Q290M9 A0A2A3E8X3 A0A084VD24 A0A2M3ZCK6 W5JRN2 A0A2P6KEH4 A0A0K8TPV1 B4LKW9 A0A182RL01 A0A336MPI5 A0A1D2MJW3 A0A2M4C4T6 B4J5I1 A0A1I8MES3 H2MII7 A0A3P9IBG9 A0A3P9LV58
A0A2J7QXL0 E2BLJ5 A0A195BMU3 A0A195F2G1 V5GDE8 A0A151J2T5 F4WIC0 E9ISY1 A0A195CM30 A0A151XBK9 A0A026WQC6 D6X268 A0A0L7QXS6 B4P4R3 A0A310SDD0 A0A1B2AJ28 B4HNC6 B4QBV9 Q7JX57 B3NLP8 Q17ED3 A0A0N0BGU2 N6T3F5 A0A1W4UZN6 A0A1Q3FJU5 A0A1Q3FIV0 A0A182QVS4 A0A182JQZ0 B3MH49 A0A2M3ZC54 B0W2N0 T1E7K1 B4GBM0 Q290M9 A0A2A3E8X3 A0A084VD24 A0A2M3ZCK6 W5JRN2 A0A2P6KEH4 A0A0K8TPV1 B4LKW9 A0A182RL01 A0A336MPI5 A0A1D2MJW3 A0A2M4C4T6 B4J5I1 A0A1I8MES3 H2MII7 A0A3P9IBG9 A0A3P9LV58
PDB
6O5B
E-value=1.50087e-08,
Score=135
Ontologies
GO
PANTHER
Topology
Subcellular location
Mitochondrion inner membrane
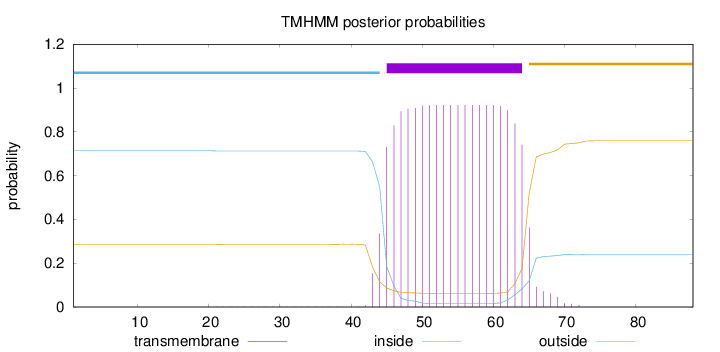
Length:
88
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.00989
Exp number, first 60 AAs:
14.93744
Total prob of N-in:
0.71412
POSSIBLE N-term signal
sequence
inside
1 - 44
TMhelix
45 - 64
outside
65 - 88
Population Genetic Test Statistics
Pi
224.272127
Theta
197.932762
Tajima's D
0.41885
CLR
0.285758
CSRT
0.490475476226189
Interpretation
Possibly Positive selection
