Pre Gene Modal
BGIBMGA005519
Annotation
PREDICTED:_protein_flightless-1_isoform_X1_[Amyelois_transitella]
Full name
Protein flightless-1
Alternative Name
Flightless-I
Location in the cell
Cytoplasmic Reliability : 2.825
Sequence
CDS
ATGCACTACGTAACGCGCTTGTACCGAGTCCACGGCGCCGGCAACAGCATCCACCTGGAGCCGGTCGCGGTGCGCGCCGCCTCGCTCGACCCGAGATACGTCTTCGTGCTCGACACCGGACTCATAATTTATCTCTGGTACGGTAAGAAAGCTAAGAACACCCTCAAGTCCAAAGCTCGTTTGTTTGCGGAGAAGATAAACAAAGAGGAGAGGAAGAACAAAGCGGAGCTGATCGCGGAACCGCCGGGGAAGGAACCCAAAAGTTTCTGGCAGATATTGGGGCACGAAGAGGATGTGCCTTTTGTACCCGAGGAGCACGTGGGCGAGTGGAGCAGCCGGGCGGTGCGGCTGTACCGCGTGGAGCTCGGCATGGGCTACCTCGAGCTGCCGCAGCCCGGGCCCGCGCCGCTGTCCCGGGGCGCGCTCGCCACTAGGAACGTTTACATCCTCGACGCGCACCTCGACGTCTTCGTCTGGTTCGGTAAGAAGTCCTCCCGGCTGGTGCGAGCAGCTGCGGTGAAGCTCGCGCAGGAGCTGTTCAACATGGTGACGCGGCCGCCGCACGCGCTCGTCACGCGTCTCCAAGAAGGGACCGAGACACAGGTGTTCAAAACGTATTTCCTGGGCTGGGACGAAATCATAGCGGTGGACTTCACGAGAACTGCAGAGTCTGTGGCGAGGACCGGGGTAGACCTCGCCAGCTGGGCCAAACAACAGGAGACTAAGGCCGATCTATCAGCGCTGTTCACGCCGAGGCAGCCGGCGATGACGGCGTCAGAAGCGAAGACCCTCGCAGACGAGTGGAACGAAGACCTGGAAGCGATGGAGGCCTTCGTGCTGGAGGGACGCCACTTCGTCCGGTTGCCCGAGAACGAACTGGGGGTCTTCTACAGCTGTGATTGCTACGTGTTCCTCTGCAGATATGTCTTGTGTCCTGATGCCGAGGAAGAGAGCGAGGGCTCCAACGTGGATCACGAGACGGAAGACGACGGTGAATCCGCCACCTGGGTCGTGTACTTCTGGCAGGGACGTAGGGCTCCCAACATGGGCTGGCTGACGTTCACCTTCGGTCTTGAGAGGAAGTTCAAGCAGCTCTGCAAGCGCCTTGAGGTCGTCCGGACGCACCAGCAGCAAGAGAGCCTCAAGTTCCTCTCGCACTTCCACAGGCAATTCGTGCTCAGAGACGGCAAGAGGAATCTTAAACCGGAGGGTCGTCTGCCCGTCGAGTTGTTCGAGCTGCGTAGTAACGGCTCCGCGCTCTGCACCAGGCTGGTCCAGGTCAAAGCGGAGGCGTCCCAACTGAACAGTGCCTTCTGCTACATTTTAAACGTACCGCTGGAAGGTAACGGGGAAACGTCGTCCGCCATCGTGTACGCGTGGATCGGGAGCAAAAGCGACCCCGACTCCGCGCGACTCATCGAGCAAATGGCCGAAGATAAGTTCAACAACCCGTGGGTCAGCCTACAGGTATTAACGGAGGGTAGCGAGCCAGATAATTTCTTCTGGGTGGCGCTCGGTGGCCGCAAGCCTTACGACACAGACGCGGACTATCTCAACTATACGAGACTGTTCCGGTGCTCAAACGAGAAAGGATACTTTACAGTCTCCGAGAAATGTACGGACTTCTGTCAGGATGATTTGGCTGATGATGATATTATGATATTGGACAACGGAGAACAGGTCTTTCTGTGGCTTGGAGCAAAATGTTCTGAAGTGGAAATAAAGCTTGCATACAAATCTGCACAGGTGTATATACAACACATGAAAACCGTACAACCAGACCGTCCAAGGAAACTGTTTTTGACACTAAAAGACAAAGAGTCACGTCGGTTCACCAAGTGCTTCCATGGCTGGGGGGATCACAAGCGGCCTCCAGAATAA
Protein
MHYVTRLYRVHGAGNSIHLEPVAVRAASLDPRYVFVLDTGLIIYLWYGKKAKNTLKSKARLFAEKINKEERKNKAELIAEPPGKEPKSFWQILGHEEDVPFVPEEHVGEWSSRAVRLYRVELGMGYLELPQPGPAPLSRGALATRNVYILDAHLDVFVWFGKKSSRLVRAAAVKLAQELFNMVTRPPHALVTRLQEGTETQVFKTYFLGWDEIIAVDFTRTAESVARTGVDLASWAKQQETKADLSALFTPRQPAMTASEAKTLADEWNEDLEAMEAFVLEGRHFVRLPENELGVFYSCDCYVFLCRYVLCPDAEEESEGSNVDHETEDDGESATWVVYFWQGRRAPNMGWLTFTFGLERKFKQLCKRLEVVRTHQQQESLKFLSHFHRQFVLRDGKRNLKPEGRLPVELFELRSNGSALCTRLVQVKAEASQLNSAFCYILNVPLEGNGETSSAIVYAWIGSKSDPDSARLIEQMAEDKFNNPWVSLQVLTEGSEPDNFFWVALGGRKPYDTDADYLNYTRLFRCSNEKGYFTVSEKCTDFCQDDLADDDIMILDNGEQVFLWLGAKCSEVEIKLAYKSAQVYIQHMKTVQPDRPRKLFLTLKDKESRRFTKCFHGWGDHKRPPE
Summary
Similarity
Belongs to the villin/gelsolin family.
Keywords
Actin-binding
Complete proteome
Developmental protein
Leucine-rich repeat
Polymorphism
Reference proteome
Repeat
Feature
chain Protein flightless-1
Uniprot
H9J7M7
A0A2H1VL32
A0A3S2P2T9
A0A2A4J2P6
A0A212ENC8
A0A2A4IZ78
+ More
A0A194PQD2 A0A2J7QXM2 A0A067QEP2 A0A2J7QXM9 A0A1Q3F6L5 A0A1Q3F6S0 E1ZXK9 E9ISY0 E2BLJ4 A0A151J2C3 A0A195F3I7 B0W2M9 A0A158NZT8 A0A0L7QXN2 A0A195BMH4 A0A026WPB4 A0A182H6N0 A0A151XBG4 Q17ED4 F4WIB9 A0A1S4F5U6 A0A195CNT6 A0A154P3I5 A0A084VD23 A0A182P0R8 A0A182UKK9 A0A182V9X0 A0A182WFQ2 A0A1B6C6V4 A0A2A3E9T0 A0A182Y6W2 A0A182NT12 A0A182IPJ9 A0A0L0BSJ3 A0A1B6I1P4 E0V9L7 A0A182RL00 A0A146KS58 D6X267 A0A182JQY9 T1P9T5 A0A336MYS3 A0A1I8MN70 A0A0A9X1H4 A0A0C9RME3 A0A310SQE9 W5JSG5 A0A0K8TKX2 A0A1I8NPL1 A0A1B0G1T6 A0A1Y1MHK3 K7IVH7 A0A0T6AZ13 A0A1A9VXJ4 A0A1L8DJ38 A0A1B0A7F4 A0A1A9Y669 A0A224XI49 A0A1J1J942 A0A0A1WWI1 A0A0K8TVK2 A0A1A9W9U5 A0A034WDM7 A0A0M9A185 A0A0P4VHI9 A0A088A2Y8 B4JIY5 B5DM61 B4L1L0 B4M353 J9JY79 B4GYE2 Q24020 B3NYD3 A0A0Q5TJ86 A0A1W4VVD9 B4I6G7 A0A3B0KM23 B3MWI7 A0A0R1EI52 B4PZ29 B4MSC1 A0A2H8TCW9 A0A2S2P6Q7 A0A226EEV5 A0A0P6FGJ4 A0A0P5N9H3 A0A0P6IBJ3 A0A0P6EZ46 A0A0P5VPY8 A0A0P6ARI5 A0A0P6EXZ2 A0A0P5ZVP3 A0A0P5TNY0
A0A194PQD2 A0A2J7QXM2 A0A067QEP2 A0A2J7QXM9 A0A1Q3F6L5 A0A1Q3F6S0 E1ZXK9 E9ISY0 E2BLJ4 A0A151J2C3 A0A195F3I7 B0W2M9 A0A158NZT8 A0A0L7QXN2 A0A195BMH4 A0A026WPB4 A0A182H6N0 A0A151XBG4 Q17ED4 F4WIB9 A0A1S4F5U6 A0A195CNT6 A0A154P3I5 A0A084VD23 A0A182P0R8 A0A182UKK9 A0A182V9X0 A0A182WFQ2 A0A1B6C6V4 A0A2A3E9T0 A0A182Y6W2 A0A182NT12 A0A182IPJ9 A0A0L0BSJ3 A0A1B6I1P4 E0V9L7 A0A182RL00 A0A146KS58 D6X267 A0A182JQY9 T1P9T5 A0A336MYS3 A0A1I8MN70 A0A0A9X1H4 A0A0C9RME3 A0A310SQE9 W5JSG5 A0A0K8TKX2 A0A1I8NPL1 A0A1B0G1T6 A0A1Y1MHK3 K7IVH7 A0A0T6AZ13 A0A1A9VXJ4 A0A1L8DJ38 A0A1B0A7F4 A0A1A9Y669 A0A224XI49 A0A1J1J942 A0A0A1WWI1 A0A0K8TVK2 A0A1A9W9U5 A0A034WDM7 A0A0M9A185 A0A0P4VHI9 A0A088A2Y8 B4JIY5 B5DM61 B4L1L0 B4M353 J9JY79 B4GYE2 Q24020 B3NYD3 A0A0Q5TJ86 A0A1W4VVD9 B4I6G7 A0A3B0KM23 B3MWI7 A0A0R1EI52 B4PZ29 B4MSC1 A0A2H8TCW9 A0A2S2P6Q7 A0A226EEV5 A0A0P6FGJ4 A0A0P5N9H3 A0A0P6IBJ3 A0A0P6EZ46 A0A0P5VPY8 A0A0P6ARI5 A0A0P6EXZ2 A0A0P5ZVP3 A0A0P5TNY0
Pubmed
19121390
22118469
26354079
24845553
20798317
21282665
+ More
21347285 24508170 30249741 26483478 17510324 21719571 24438588 25244985 26108605 20566863 26823975 18362917 19820115 25315136 25401762 20920257 23761445 26369729 28004739 20075255 25830018 25348373 27129103 17994087 15632085 8248259 8582612 9520435 10731132 12537572 10731138 18057021 17550304
21347285 24508170 30249741 26483478 17510324 21719571 24438588 25244985 26108605 20566863 26823975 18362917 19820115 25315136 25401762 20920257 23761445 26369729 28004739 20075255 25830018 25348373 27129103 17994087 15632085 8248259 8582612 9520435 10731132 12537572 10731138 18057021 17550304
EMBL
BABH01014058
BABH01014059
BABH01014060
BABH01014061
ODYU01003149
SOQ41533.1
+ More
RSAL01000041 RVE50907.1 NWSH01003929 PCG65672.1 AGBW02013681 OWR42979.1 NWSH01005084 PCG64452.1 KQ459595 KPI95656.1 NEVH01009378 PNF33329.1 KK853751 KDQ85824.1 PNF33328.1 GFDL01011858 JAV23187.1 GFDL01011789 JAV23256.1 GL435066 EFN74092.1 GL765434 EFZ16387.1 GL449033 EFN83431.1 KQ980404 KYN16191.1 KQ981856 KYN34649.1 DS231827 EDS29337.1 ADTU01005002 ADTU01005003 KQ414704 KOC63357.1 KQ976440 KYM86486.1 KK107139 QOIP01000003 EZA57897.1 RLU25165.1 JXUM01114411 JXUM01114412 KQ565783 KXJ70670.1 KQ982330 KYQ57679.1 CH477283 EAT44842.1 GL888175 EGI66050.1 KQ977580 KYN01764.1 KQ434809 KZC06476.1 ATLV01011135 KE524643 KFB35867.1 GEDC01028164 JAS09134.1 KZ288322 PBC28234.1 JRES01001421 KNC23050.1 GECU01026858 JAS80848.1 DS234994 EEB10073.1 GDHC01019305 JAP99323.1 KQ971371 EFA10220.1 KA645516 AFP60145.1 UFQS01001564 UFQT01001564 SSX11461.1 SSX31028.1 GBHO01029745 JAG13859.1 GBYB01008151 GBYB01011973 JAG77918.1 JAG81740.1 KQ761039 OAD58387.1 ADMH02000297 ETN67066.1 GDAI01002817 JAI14786.1 CCAG010006899 GEZM01034411 JAV83815.1 LJIG01022475 KRT80377.1 GFDF01007699 JAV06385.1 GFTR01008707 JAW07719.1 CVRI01000072 CRL07521.1 GBXI01011437 JAD02855.1 GDHF01034011 GDHF01033377 GDHF01033369 GDHF01028190 GDHF01017032 GDHF01000295 JAI18303.1 JAI18937.1 JAI18945.1 JAI24124.1 JAI35282.1 JAI52019.1 GAKP01007059 JAC51893.1 KQ435767 KOX75280.1 GDKW01002650 JAI53945.1 CH916370 EDV99549.1 CH379064 EDY72578.1 CH933810 EDW07646.1 CH940651 EDW65228.2 ABLF02016317 CH479197 EDW27798.1 U01182 AF017777 AE014298 AF132184 CH954180 EDV47612.1 KQS30573.1 KQS30574.1 CH480823 EDW56373.1 OUUW01000011 SPP86886.1 CH902625 EDV34972.2 CM000162 KRK07105.1 EDX03090.1 CH963851 EDW75010.1 GFXV01000134 MBW11939.1 GGMR01012476 MBY25095.1 LNIX01000004 OXA56072.1 GDIQ01064587 JAN30150.1 GDIQ01145317 GDIQ01039705 JAL06409.1 GDIQ01006918 JAN87819.1 GDIQ01056541 JAN38196.1 GDIP01097075 JAM06640.1 GDIP01025550 JAM78165.1 GDIQ01056539 JAN38198.1 GDIP01037891 JAM65824.1 GDIP01124941 JAL78773.1
RSAL01000041 RVE50907.1 NWSH01003929 PCG65672.1 AGBW02013681 OWR42979.1 NWSH01005084 PCG64452.1 KQ459595 KPI95656.1 NEVH01009378 PNF33329.1 KK853751 KDQ85824.1 PNF33328.1 GFDL01011858 JAV23187.1 GFDL01011789 JAV23256.1 GL435066 EFN74092.1 GL765434 EFZ16387.1 GL449033 EFN83431.1 KQ980404 KYN16191.1 KQ981856 KYN34649.1 DS231827 EDS29337.1 ADTU01005002 ADTU01005003 KQ414704 KOC63357.1 KQ976440 KYM86486.1 KK107139 QOIP01000003 EZA57897.1 RLU25165.1 JXUM01114411 JXUM01114412 KQ565783 KXJ70670.1 KQ982330 KYQ57679.1 CH477283 EAT44842.1 GL888175 EGI66050.1 KQ977580 KYN01764.1 KQ434809 KZC06476.1 ATLV01011135 KE524643 KFB35867.1 GEDC01028164 JAS09134.1 KZ288322 PBC28234.1 JRES01001421 KNC23050.1 GECU01026858 JAS80848.1 DS234994 EEB10073.1 GDHC01019305 JAP99323.1 KQ971371 EFA10220.1 KA645516 AFP60145.1 UFQS01001564 UFQT01001564 SSX11461.1 SSX31028.1 GBHO01029745 JAG13859.1 GBYB01008151 GBYB01011973 JAG77918.1 JAG81740.1 KQ761039 OAD58387.1 ADMH02000297 ETN67066.1 GDAI01002817 JAI14786.1 CCAG010006899 GEZM01034411 JAV83815.1 LJIG01022475 KRT80377.1 GFDF01007699 JAV06385.1 GFTR01008707 JAW07719.1 CVRI01000072 CRL07521.1 GBXI01011437 JAD02855.1 GDHF01034011 GDHF01033377 GDHF01033369 GDHF01028190 GDHF01017032 GDHF01000295 JAI18303.1 JAI18937.1 JAI18945.1 JAI24124.1 JAI35282.1 JAI52019.1 GAKP01007059 JAC51893.1 KQ435767 KOX75280.1 GDKW01002650 JAI53945.1 CH916370 EDV99549.1 CH379064 EDY72578.1 CH933810 EDW07646.1 CH940651 EDW65228.2 ABLF02016317 CH479197 EDW27798.1 U01182 AF017777 AE014298 AF132184 CH954180 EDV47612.1 KQS30573.1 KQS30574.1 CH480823 EDW56373.1 OUUW01000011 SPP86886.1 CH902625 EDV34972.2 CM000162 KRK07105.1 EDX03090.1 CH963851 EDW75010.1 GFXV01000134 MBW11939.1 GGMR01012476 MBY25095.1 LNIX01000004 OXA56072.1 GDIQ01064587 JAN30150.1 GDIQ01145317 GDIQ01039705 JAL06409.1 GDIQ01006918 JAN87819.1 GDIQ01056541 JAN38196.1 GDIP01097075 JAM06640.1 GDIP01025550 JAM78165.1 GDIQ01056539 JAN38198.1 GDIP01037891 JAM65824.1 GDIP01124941 JAL78773.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053268
UP000235965
+ More
UP000027135 UP000000311 UP000008237 UP000078492 UP000078541 UP000002320 UP000005205 UP000053825 UP000078540 UP000053097 UP000279307 UP000069940 UP000249989 UP000075809 UP000008820 UP000007755 UP000078542 UP000076502 UP000030765 UP000075885 UP000075902 UP000075903 UP000075920 UP000242457 UP000076408 UP000075884 UP000075880 UP000037069 UP000009046 UP000075900 UP000007266 UP000075881 UP000095301 UP000000673 UP000095300 UP000092444 UP000002358 UP000078200 UP000092445 UP000092443 UP000183832 UP000091820 UP000053105 UP000005203 UP000001070 UP000001819 UP000009192 UP000008792 UP000007819 UP000008744 UP000000803 UP000008711 UP000192221 UP000001292 UP000268350 UP000007801 UP000002282 UP000007798 UP000198287
UP000027135 UP000000311 UP000008237 UP000078492 UP000078541 UP000002320 UP000005205 UP000053825 UP000078540 UP000053097 UP000279307 UP000069940 UP000249989 UP000075809 UP000008820 UP000007755 UP000078542 UP000076502 UP000030765 UP000075885 UP000075902 UP000075903 UP000075920 UP000242457 UP000076408 UP000075884 UP000075880 UP000037069 UP000009046 UP000075900 UP000007266 UP000075881 UP000095301 UP000000673 UP000095300 UP000092444 UP000002358 UP000078200 UP000092445 UP000092443 UP000183832 UP000091820 UP000053105 UP000005203 UP000001070 UP000001819 UP000009192 UP000008792 UP000007819 UP000008744 UP000000803 UP000008711 UP000192221 UP000001292 UP000268350 UP000007801 UP000002282 UP000007798 UP000198287
Interpro
Gene 3D
ProteinModelPortal
H9J7M7
A0A2H1VL32
A0A3S2P2T9
A0A2A4J2P6
A0A212ENC8
A0A2A4IZ78
+ More
A0A194PQD2 A0A2J7QXM2 A0A067QEP2 A0A2J7QXM9 A0A1Q3F6L5 A0A1Q3F6S0 E1ZXK9 E9ISY0 E2BLJ4 A0A151J2C3 A0A195F3I7 B0W2M9 A0A158NZT8 A0A0L7QXN2 A0A195BMH4 A0A026WPB4 A0A182H6N0 A0A151XBG4 Q17ED4 F4WIB9 A0A1S4F5U6 A0A195CNT6 A0A154P3I5 A0A084VD23 A0A182P0R8 A0A182UKK9 A0A182V9X0 A0A182WFQ2 A0A1B6C6V4 A0A2A3E9T0 A0A182Y6W2 A0A182NT12 A0A182IPJ9 A0A0L0BSJ3 A0A1B6I1P4 E0V9L7 A0A182RL00 A0A146KS58 D6X267 A0A182JQY9 T1P9T5 A0A336MYS3 A0A1I8MN70 A0A0A9X1H4 A0A0C9RME3 A0A310SQE9 W5JSG5 A0A0K8TKX2 A0A1I8NPL1 A0A1B0G1T6 A0A1Y1MHK3 K7IVH7 A0A0T6AZ13 A0A1A9VXJ4 A0A1L8DJ38 A0A1B0A7F4 A0A1A9Y669 A0A224XI49 A0A1J1J942 A0A0A1WWI1 A0A0K8TVK2 A0A1A9W9U5 A0A034WDM7 A0A0M9A185 A0A0P4VHI9 A0A088A2Y8 B4JIY5 B5DM61 B4L1L0 B4M353 J9JY79 B4GYE2 Q24020 B3NYD3 A0A0Q5TJ86 A0A1W4VVD9 B4I6G7 A0A3B0KM23 B3MWI7 A0A0R1EI52 B4PZ29 B4MSC1 A0A2H8TCW9 A0A2S2P6Q7 A0A226EEV5 A0A0P6FGJ4 A0A0P5N9H3 A0A0P6IBJ3 A0A0P6EZ46 A0A0P5VPY8 A0A0P6ARI5 A0A0P6EXZ2 A0A0P5ZVP3 A0A0P5TNY0
A0A194PQD2 A0A2J7QXM2 A0A067QEP2 A0A2J7QXM9 A0A1Q3F6L5 A0A1Q3F6S0 E1ZXK9 E9ISY0 E2BLJ4 A0A151J2C3 A0A195F3I7 B0W2M9 A0A158NZT8 A0A0L7QXN2 A0A195BMH4 A0A026WPB4 A0A182H6N0 A0A151XBG4 Q17ED4 F4WIB9 A0A1S4F5U6 A0A195CNT6 A0A154P3I5 A0A084VD23 A0A182P0R8 A0A182UKK9 A0A182V9X0 A0A182WFQ2 A0A1B6C6V4 A0A2A3E9T0 A0A182Y6W2 A0A182NT12 A0A182IPJ9 A0A0L0BSJ3 A0A1B6I1P4 E0V9L7 A0A182RL00 A0A146KS58 D6X267 A0A182JQY9 T1P9T5 A0A336MYS3 A0A1I8MN70 A0A0A9X1H4 A0A0C9RME3 A0A310SQE9 W5JSG5 A0A0K8TKX2 A0A1I8NPL1 A0A1B0G1T6 A0A1Y1MHK3 K7IVH7 A0A0T6AZ13 A0A1A9VXJ4 A0A1L8DJ38 A0A1B0A7F4 A0A1A9Y669 A0A224XI49 A0A1J1J942 A0A0A1WWI1 A0A0K8TVK2 A0A1A9W9U5 A0A034WDM7 A0A0M9A185 A0A0P4VHI9 A0A088A2Y8 B4JIY5 B5DM61 B4L1L0 B4M353 J9JY79 B4GYE2 Q24020 B3NYD3 A0A0Q5TJ86 A0A1W4VVD9 B4I6G7 A0A3B0KM23 B3MWI7 A0A0R1EI52 B4PZ29 B4MSC1 A0A2H8TCW9 A0A2S2P6Q7 A0A226EEV5 A0A0P6FGJ4 A0A0P5N9H3 A0A0P6IBJ3 A0A0P6EZ46 A0A0P5VPY8 A0A0P6ARI5 A0A0P6EXZ2 A0A0P5ZVP3 A0A0P5TNY0
PDB
3FFN
E-value=2.16783e-36,
Score=384
Ontologies
GO
PANTHER
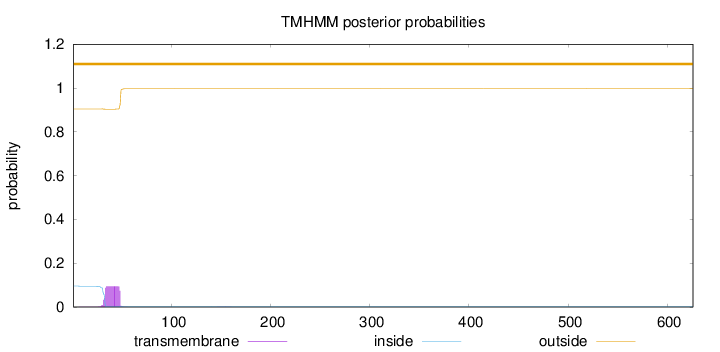
Topology
Length:
626
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.6128
Exp number, first 60 AAs:
1.60843
Total prob of N-in:
0.09555
outside
1 - 626
Population Genetic Test Statistics
Pi
226.828743
Theta
158.817938
Tajima's D
-2.646282
CLR
1172.589524
CSRT
0.000149992500374981
Interpretation
Possibly Positive selection
