Gene
KWMTBOMO04668 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005519
Annotation
PREDICTED:_mitochondrial_import_receptor_subunit_TOM22_homolog_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 1.918
Sequence
CDS
ATGCTTATGGATTTGACCGATGAATTAGAATTAAGCGATAGCGGAATGGAGTCTTTGACTACGAGCAAAGATGATACCCCTGAAAGAAGGCCTGAACAATATTTCATAACCCCCTCGGTGGGGTCATCGCCCACTGCTGGATCTCCTAGTGGACCTTTGAAAGAGTACGACGATGAACCAGATGAAACTCTTTCTGAAAGATTGTGGGGTCTGACCGAAATGTTCCCAGAGTGTGTAAGGAATGGCACATACACAGTTACAACAAATACATGGTCTGGTATCAAAGGCCTGTATGGTCTATCACGCTCAGTAATGTGGGTTGTTGCCAGTTCATCGGTAATATTGTTTGCTCCTGTCATATTTGAGGTAGAAAGAGCTCAAGTAGCCGAAATGGAAAAGTCACAACAAAAACAGGTGTTATTAGGGACAAACACAGCTATGGCAGGACCCATGCCTGCGATGCCACCAATGCCACGATAA
Protein
MLMDLTDELELSDSGMESLTTSKDDTPERRPEQYFITPSVGSSPTAGSPSGPLKEYDDEPDETLSERLWGLTEMFPECVRNGTYTVTTNTWSGIKGLYGLSRSVMWVVASSSVILFAPVIFEVERAQVAEMEKSQQKQVLLGTNTAMAGPMPAMPPMPR
Summary
Uniprot
H9J7M7
A0A3S2NLR7
A0A2H1WQ16
S4PYD4
A0A1E1WVD6
A0A212ENB6
+ More
A0A194PSC1 A0A0L7LV22 A0A194QYC9 D6X290 B3M7Z4 B3NFZ6 B4PH77 A0A0J9UE36 Q9VZL1 W8AZI4 A0A0K8UH19 A0A034V3U3 A0A336MYG2 A0A1W4W1V8 A0A1B0FB35 A0A0K8TS13 A0A0M5J691 A0A1J1IXJ8 A0A1B0AI18 A0A0A9Y9P7 A0A3B0KAT0 B4KXU0 D3TMG8 A0A1A9VYS1 A0A1B0BPW6 A0A0L0BQF5 A0A0P4VXR0 R4FKA1 A0A069DP57 A0A224XP97 U5EYK4 A0A1I8MLE8 B4MGL6 Q29EZ3 A0A182KW53 B4N6C8 B4H5Q3 A0A1L8EAU8 B4IZ32 A0A1L8EAH3 A0A0V0G9K1 R4WKA9 T1PC94 A0A1I8PEI4 A0A1B6MRY6 A0A1L8E171 J3JXK4
A0A194PSC1 A0A0L7LV22 A0A194QYC9 D6X290 B3M7Z4 B3NFZ6 B4PH77 A0A0J9UE36 Q9VZL1 W8AZI4 A0A0K8UH19 A0A034V3U3 A0A336MYG2 A0A1W4W1V8 A0A1B0FB35 A0A0K8TS13 A0A0M5J691 A0A1J1IXJ8 A0A1B0AI18 A0A0A9Y9P7 A0A3B0KAT0 B4KXU0 D3TMG8 A0A1A9VYS1 A0A1B0BPW6 A0A0L0BQF5 A0A0P4VXR0 R4FKA1 A0A069DP57 A0A224XP97 U5EYK4 A0A1I8MLE8 B4MGL6 Q29EZ3 A0A182KW53 B4N6C8 B4H5Q3 A0A1L8EAU8 B4IZ32 A0A1L8EAH3 A0A0V0G9K1 R4WKA9 T1PC94 A0A1I8PEI4 A0A1B6MRY6 A0A1L8E171 J3JXK4
Pubmed
19121390
23622113
22118469
26354079
26227816
18362917
+ More
19820115 17994087 17550304 22936249 10978288 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 25348373 26369729 25401762 26823975 20353571 26108605 27129103 26334808 25315136 15632085 20966253 23691247 22516182 23537049
19820115 17994087 17550304 22936249 10978288 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 25348373 26369729 25401762 26823975 20353571 26108605 27129103 26334808 25315136 15632085 20966253 23691247 22516182 23537049
EMBL
BABH01014058
BABH01014059
BABH01014060
BABH01014061
RSAL01000041
RVE50906.1
+ More
ODYU01010204 SOQ55165.1 GAIX01003483 JAA89077.1 GDQN01000122 JAT90932.1 AGBW02013681 OWR42978.1 KQ459595 KPI95654.1 JTDY01000040 KOB79234.1 KQ461154 KPJ08581.1 KQ971371 EFA10229.1 CH902618 EDV39902.1 CH954178 EDV50825.2 CM000159 EDW93314.2 CM002912 KMY97440.1 AE014296 AF241222 AY122211 AAF47809.1 AAF67180.1 AAM52723.1 GAMC01012310 JAB94245.1 GDHF01026456 JAI25858.1 GAKP01022507 JAC36445.1 UFQT01000200 UFQT01003214 SSX21614.1 SSX34735.1 CCAG010007616 CCAG010007617 CCAG010007618 GDAI01000657 JAI16946.1 CP012525 ALC44818.1 CVRI01000063 CRL04442.1 GBHO01014700 GBRD01010803 GDHC01020038 JAG28904.1 JAG55021.1 JAP98590.1 OUUW01000009 SPP85240.1 CH933809 EDW17612.1 EZ422620 ADD18896.1 JXJN01018245 JRES01001517 KNC22320.1 GDKW01002209 JAI54386.1 ACPB03025641 GAHY01001948 JAA75562.1 GBGD01003433 JAC85456.1 GFTR01002119 JAW14307.1 GANO01000298 JAB59573.1 CH940682 EDW57082.1 CH379070 EAL29916.1 CH964154 EDW79917.1 CH479211 EDW33105.1 GFDG01003106 JAV15693.1 CH916366 EDV95554.1 GFDG01003194 JAV15605.1 GECL01001274 JAP04850.1 AK418015 BAN21230.1 KA646309 AFP60938.1 GEBQ01001299 JAT38678.1 GFDF01001637 JAV12447.1 APGK01042895 BT127973 KB741009 KB632250 AEE62935.1 ENN75602.1 ERL90596.1
ODYU01010204 SOQ55165.1 GAIX01003483 JAA89077.1 GDQN01000122 JAT90932.1 AGBW02013681 OWR42978.1 KQ459595 KPI95654.1 JTDY01000040 KOB79234.1 KQ461154 KPJ08581.1 KQ971371 EFA10229.1 CH902618 EDV39902.1 CH954178 EDV50825.2 CM000159 EDW93314.2 CM002912 KMY97440.1 AE014296 AF241222 AY122211 AAF47809.1 AAF67180.1 AAM52723.1 GAMC01012310 JAB94245.1 GDHF01026456 JAI25858.1 GAKP01022507 JAC36445.1 UFQT01000200 UFQT01003214 SSX21614.1 SSX34735.1 CCAG010007616 CCAG010007617 CCAG010007618 GDAI01000657 JAI16946.1 CP012525 ALC44818.1 CVRI01000063 CRL04442.1 GBHO01014700 GBRD01010803 GDHC01020038 JAG28904.1 JAG55021.1 JAP98590.1 OUUW01000009 SPP85240.1 CH933809 EDW17612.1 EZ422620 ADD18896.1 JXJN01018245 JRES01001517 KNC22320.1 GDKW01002209 JAI54386.1 ACPB03025641 GAHY01001948 JAA75562.1 GBGD01003433 JAC85456.1 GFTR01002119 JAW14307.1 GANO01000298 JAB59573.1 CH940682 EDW57082.1 CH379070 EAL29916.1 CH964154 EDW79917.1 CH479211 EDW33105.1 GFDG01003106 JAV15693.1 CH916366 EDV95554.1 GFDG01003194 JAV15605.1 GECL01001274 JAP04850.1 AK418015 BAN21230.1 KA646309 AFP60938.1 GEBQ01001299 JAT38678.1 GFDF01001637 JAV12447.1 APGK01042895 BT127973 KB741009 KB632250 AEE62935.1 ENN75602.1 ERL90596.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000037510
UP000053240
+ More
UP000007266 UP000007801 UP000008711 UP000002282 UP000000803 UP000192221 UP000092444 UP000092553 UP000183832 UP000092445 UP000268350 UP000009192 UP000078200 UP000092460 UP000037069 UP000015103 UP000095301 UP000008792 UP000001819 UP000075882 UP000007798 UP000008744 UP000001070 UP000095300 UP000019118 UP000030742
UP000007266 UP000007801 UP000008711 UP000002282 UP000000803 UP000192221 UP000092444 UP000092553 UP000183832 UP000092445 UP000268350 UP000009192 UP000078200 UP000092460 UP000037069 UP000015103 UP000095301 UP000008792 UP000001819 UP000075882 UP000007798 UP000008744 UP000001070 UP000095300 UP000019118 UP000030742
Interpro
Gene 3D
ProteinModelPortal
H9J7M7
A0A3S2NLR7
A0A2H1WQ16
S4PYD4
A0A1E1WVD6
A0A212ENB6
+ More
A0A194PSC1 A0A0L7LV22 A0A194QYC9 D6X290 B3M7Z4 B3NFZ6 B4PH77 A0A0J9UE36 Q9VZL1 W8AZI4 A0A0K8UH19 A0A034V3U3 A0A336MYG2 A0A1W4W1V8 A0A1B0FB35 A0A0K8TS13 A0A0M5J691 A0A1J1IXJ8 A0A1B0AI18 A0A0A9Y9P7 A0A3B0KAT0 B4KXU0 D3TMG8 A0A1A9VYS1 A0A1B0BPW6 A0A0L0BQF5 A0A0P4VXR0 R4FKA1 A0A069DP57 A0A224XP97 U5EYK4 A0A1I8MLE8 B4MGL6 Q29EZ3 A0A182KW53 B4N6C8 B4H5Q3 A0A1L8EAU8 B4IZ32 A0A1L8EAH3 A0A0V0G9K1 R4WKA9 T1PC94 A0A1I8PEI4 A0A1B6MRY6 A0A1L8E171 J3JXK4
A0A194PSC1 A0A0L7LV22 A0A194QYC9 D6X290 B3M7Z4 B3NFZ6 B4PH77 A0A0J9UE36 Q9VZL1 W8AZI4 A0A0K8UH19 A0A034V3U3 A0A336MYG2 A0A1W4W1V8 A0A1B0FB35 A0A0K8TS13 A0A0M5J691 A0A1J1IXJ8 A0A1B0AI18 A0A0A9Y9P7 A0A3B0KAT0 B4KXU0 D3TMG8 A0A1A9VYS1 A0A1B0BPW6 A0A0L0BQF5 A0A0P4VXR0 R4FKA1 A0A069DP57 A0A224XP97 U5EYK4 A0A1I8MLE8 B4MGL6 Q29EZ3 A0A182KW53 B4N6C8 B4H5Q3 A0A1L8EAU8 B4IZ32 A0A1L8EAH3 A0A0V0G9K1 R4WKA9 T1PC94 A0A1I8PEI4 A0A1B6MRY6 A0A1L8E171 J3JXK4
Ontologies
GO
PANTHER
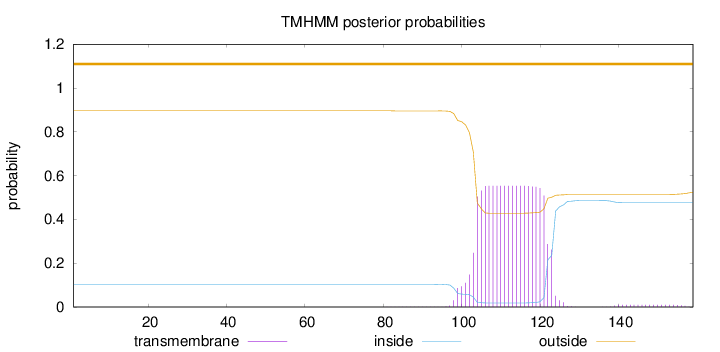
Topology
Length:
159
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
11.40039
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.10416
outside
1 - 159
Population Genetic Test Statistics
Pi
126.86979
Theta
148.380608
Tajima's D
-0.433369
CLR
254.055023
CSRT
0.250987450627469
Interpretation
Uncertain
