Gene
KWMTBOMO04654
Pre Gene Modal
BGIBMGA005300
Annotation
PREDICTED:_gamma-glutamyltranspeptidase_1_isoform_X2_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.393
Sequence
CDS
ATGACGATTCGCAGCATGACAGTTACTGAGGCGAGAGATGCGACTCGAAAACATTGCTGTGCAAGGCCTTATAAAACGAAGACCAAGTTGACAATTGCGGGGCTCGTCGTCCTCGTTATTTCATTAGGACTGGCCGGTTACTTCATAGGCGGTGCAGACTACAAGGCGGCACATCGTTTGGCGAATCCGCCGGACCCTATAGAACCATTGAAACCTTCAGCATCAACTTTACACGTGTTTCAAAAAGCGGCTGTTTGCACGGACTCGCCGCAATGCTCACAGATCGGCAGAGAAATCCTGCTGAAGAATGGCTCAGCGGTGGATGCAGCGATCGCCGCCATGTTCTGCAACAGTCTACGGAATCAGCAGAGCATGGGTCTGGGGGGTGGATTCTTCATGACCGTTTATATTAAAGAAGAGGAGAAGGCGTACACCGTGATAGCCAGGGAGAAGGCTCCTGCCGCCGCTACTGTTGACATGTTCAACGGAGACTTCACAAAGGCTTCTAAAGGTCCGCTAGCCGTTGGAGTTCCCGGGGAACTACGTGGTATGTGGGCAGCACACAGGCGCTGGGGCAAACTACCCTGGGACAGTTTAATCGAGCCAACCCTGCAGATATGCAACGAGGGCTTCCCACTTTCCAAACCTATGTACGACGCCTTGGACAAAGCACGAGCCTACGTGAAGAATGATGTCAATTTGGCGAAAATGTACTACAGTTCTTCAATGGAGAGGTTTCATAGACCAGGAACAATCGTGAAGCCAAGTGAAGCCCTTTGCAATACGTTCAAGAGGATAGCCAAGAATGGCGGAGACGACCTGTACAATGGCACTTTGGCTGAAGACTTCGTTGAAGATCTGAAAAGGATCGGAAGTATTATCACCGCTGAGGACATGAGAAATTATGAAGCTAAAATAACAGAGCCCATGACAGTGCCTCTGGGCAACGGTGATACTCTATACACCCCACCGCCGCCCAGCAGTGGGCTTATTCTCGTGAATATACTTAACATCTTGAGTGGGTACGGCTTCAATGCGGCCAGCATCAATAGCACTGAAAACATGATACTCACTTATCATAGAATAATAGAATCATTTAAATACGCGTACGCGACCCGCACTAAATTGGGCGATATGGAGTTCCTGGATTTGAAAGAGTTGATAAATAACGTTACGAAGCCGGAGTATGGCGCGGAAATCCGTATGAAAATCAACGAATCATCGACAAGTAACGATACTGCAACGTACGGAGCTGAAGATTATAATCCTCCTGATTCTGGGACCGCTCACATATCTATCATAGGGACAAATGGGGACGCTGTTTCCGTAACCAGTTCCATCAACTTTTACTACGGTGCTGGTTTTTCGACCCTCAACACCGGTATCGTGATGAACAACGTAATGGACGACTTCTCATCACCTGGAATCATTAATTATTTCGGCTTAAAGCCTTCGCCCGCTAACTTTATAGCGCCAGGCAAAAGGCCGTTGTCTTCAATGAGTCCAAGCATTATTGTAGATAAGAATGGAAATGCGAAGATGGTGATCGGTGCCTCCGGAGGAACCAAAATCACAACGGGTATTGCTTTGGTGACAATGGGCAAGCTATGGTTCGGTCAGACAATCAAAGAAGCCGTGGATAGCCCCCGAATACACCACCAATTGTTTCCTATGAACATCGAATATGAGTACGGAATTATTGAGGACGTAGTGAAAGGACTAAGGGAGAAAGGTCACGGCATGATTCGGTACAGAGATAGGGGGTCAGTGATATGTGCCCTCTTTAGAAACAAAACGGGGATTTACGCAAACGCTGACTTCAGGAAAGGAGGGGACGTCGCCGGTATGGACTGA
Protein
MTIRSMTVTEARDATRKHCCARPYKTKTKLTIAGLVVLVISLGLAGYFIGGADYKAAHRLANPPDPIEPLKPSASTLHVFQKAAVCTDSPQCSQIGREILLKNGSAVDAAIAAMFCNSLRNQQSMGLGGGFFMTVYIKEEEKAYTVIAREKAPAAATVDMFNGDFTKASKGPLAVGVPGELRGMWAAHRRWGKLPWDSLIEPTLQICNEGFPLSKPMYDALDKARAYVKNDVNLAKMYYSSSMERFHRPGTIVKPSEALCNTFKRIAKNGGDDLYNGTLAEDFVEDLKRIGSIITAEDMRNYEAKITEPMTVPLGNGDTLYTPPPPSSGLILVNILNILSGYGFNAASINSTENMILTYHRIIESFKYAYATRTKLGDMEFLDLKELINNVTKPEYGAEIRMKINESSTSNDTATYGAEDYNPPDSGTAHISIIGTNGDAVSVTSSINFYYGAGFSTLNTGIVMNNVMDDFSSPGIINYFGLKPSPANFIAPGKRPLSSMSPSIIVDKNGNAKMVIGASGGTKITTGIALVTMGKLWFGQTIKEAVDSPRIHHQLFPMNIEYEYGIIEDVVKGLREKGHGMIRYRDRGSVICALFRNKTGIYANADFRKGGDVAGMD
Summary
Uniprot
H9J708
A0A2H1VR65
A0A212FL94
A0A3S2M4R0
A0A2A4J4U1
A0A0L7KTC2
+ More
A0A194PRR0 A0A0L7KS97 A0A1W4W5B0 A0A1W4WGT8 A0A194QUB0 A0A1W7R8L0 A0A1B6DXA4 A0A084WFD9 A0A1B6J9Q8 A0A1B6C9Z6 A0A182LLY2 A0A1S4GA40 Q7Q4V8 A0A182WZ83 A0A182V524 A0A182HMQ2 A0A1Y1NPC6 A0A182THA0 A0A182ITF2 W5JUK0 A0A182N129 A0A182FJI4 A0A154PFR4 A0A1L8E287 A0A1L8E2T4 A0A182R242 A0A182QSP5 A0A0L7QZ87 A0A182XYC3 A0A182K915 B0XAA5 A0A023F196 A0A088AQ38 K7IVN9 A0A2J7QZH2 A0A3L8DL94 U5ET38 A0A195FXI2 A0A182PUB7 A0A0M8ZW98 A0A026X1J2 F4X7W8 E9JAF8 A0A2A3EJU3 D6WXU9 A0A232EQX8 E2A4N2 A0A151IVG1 A0A158NUF5 A0A195BY63 U4UJX4 N6TDX5 A0A182T6J5 E2BFT6 A0A067R874 A0A195BTE4 A0A310SUV0 A0A1I8Q012 A0A1B0EW75 A0A182LRX3 A0A1I8Q008 A0A1I8PZY1 A0A0A1WWD9 A0A0A1WKZ4 A0A0A1XBJ8 A0A0C9PIR3 W8AVQ0 A0A182VVH1 T1P7W8 A0A1I8N6G7 A0A1B0CWU0 V4BXA9 W8BI70 W8BUZ8 A0A034VHE3 A0A2R7WF77 A0A2J7QZE7 A0A034VJQ4 A0A0K8UQ34 A0A0P5TFY2 A0A0P5N4Y6 A0A0P6EPA7 A0A0K8UNY1 A0A0P4W215 A0A0P6GP16 A0A0K8VC77 A0A0P5EFG0 A0A0P6ETL0 A0A0P6A4P7 A0A0P6GRV0 A0A0P5GVJ6 E9H1T9 A0A164NFS1 A0A0P6HKY5
A0A194PRR0 A0A0L7KS97 A0A1W4W5B0 A0A1W4WGT8 A0A194QUB0 A0A1W7R8L0 A0A1B6DXA4 A0A084WFD9 A0A1B6J9Q8 A0A1B6C9Z6 A0A182LLY2 A0A1S4GA40 Q7Q4V8 A0A182WZ83 A0A182V524 A0A182HMQ2 A0A1Y1NPC6 A0A182THA0 A0A182ITF2 W5JUK0 A0A182N129 A0A182FJI4 A0A154PFR4 A0A1L8E287 A0A1L8E2T4 A0A182R242 A0A182QSP5 A0A0L7QZ87 A0A182XYC3 A0A182K915 B0XAA5 A0A023F196 A0A088AQ38 K7IVN9 A0A2J7QZH2 A0A3L8DL94 U5ET38 A0A195FXI2 A0A182PUB7 A0A0M8ZW98 A0A026X1J2 F4X7W8 E9JAF8 A0A2A3EJU3 D6WXU9 A0A232EQX8 E2A4N2 A0A151IVG1 A0A158NUF5 A0A195BY63 U4UJX4 N6TDX5 A0A182T6J5 E2BFT6 A0A067R874 A0A195BTE4 A0A310SUV0 A0A1I8Q012 A0A1B0EW75 A0A182LRX3 A0A1I8Q008 A0A1I8PZY1 A0A0A1WWD9 A0A0A1WKZ4 A0A0A1XBJ8 A0A0C9PIR3 W8AVQ0 A0A182VVH1 T1P7W8 A0A1I8N6G7 A0A1B0CWU0 V4BXA9 W8BI70 W8BUZ8 A0A034VHE3 A0A2R7WF77 A0A2J7QZE7 A0A034VJQ4 A0A0K8UQ34 A0A0P5TFY2 A0A0P5N4Y6 A0A0P6EPA7 A0A0K8UNY1 A0A0P4W215 A0A0P6GP16 A0A0K8VC77 A0A0P5EFG0 A0A0P6ETL0 A0A0P6A4P7 A0A0P6GRV0 A0A0P5GVJ6 E9H1T9 A0A164NFS1 A0A0P6HKY5
Pubmed
EMBL
BABH01014021
ODYU01003783
SOQ42972.1
AGBW02007817
OWR54514.1
RSAL01000041
+ More
RVE50892.1 NWSH01003215 PCG66756.1 JTDY01006025 KOB66355.1 KQ459595 KPI95638.1 JTDY01006550 KOB65899.1 KQ461154 KPJ08565.1 GEHC01000166 JAV47479.1 GEDC01007011 JAS30287.1 ATLV01023334 KE525342 KFB48933.1 GECU01011782 JAS95924.1 GEDC01027228 JAS10070.1 AAAB01008963 EAA12089.5 APCN01004856 GEZM01001268 GEZM01001267 JAV98007.1 ADMH02000457 ETN66414.1 KQ434893 KZC10657.1 GFDF01001267 JAV12817.1 GFDF01001279 JAV12805.1 AXCN02002173 KQ414685 KOC63852.1 DS232576 EDS43602.1 GBBI01003926 JAC14786.1 NEVH01009069 PNF33972.1 QOIP01000006 RLU21214.1 GANO01002943 JAB56928.1 KQ981193 KYN45136.1 KQ435840 KOX71346.1 KK107039 EZA61886.1 GL888900 EGI57454.1 GL770322 EFZ10192.1 KZ288232 PBC31469.1 KQ971362 EFA08906.2 NNAY01002686 OXU20775.1 GL436710 EFN71634.1 KQ980900 KYN11703.1 ADTU01026413 ADTU01026414 KQ978501 KYM93599.1 KB632356 ERL93407.1 APGK01041542 APGK01041543 KB740994 ENN75953.1 GL448039 EFN85474.1 KK852817 KDR15749.1 KQ976417 KYM89948.1 KQ760221 OAD61360.1 AJVK01012183 AJVK01012184 AJVK01012185 AJVK01012186 AXCM01003938 GBXI01011492 JAD02800.1 GBXI01015214 JAC99077.1 GBXI01010591 GBXI01009712 GBXI01006454 JAD03701.1 JAD04580.1 JAD07838.1 GBYB01000838 JAG70605.1 GAMC01013620 GAMC01013618 JAB92935.1 KA644642 AFP59271.1 AJWK01032855 KB201890 ESO93724.1 GAMC01013619 JAB92936.1 GAMC01013621 JAB92934.1 GAKP01016266 JAC42686.1 KK854575 PTY17025.1 PNF33971.1 GAKP01016263 JAC42689.1 GDHF01023525 JAI28789.1 GDIQ01092085 JAL59641.1 GDIQ01147228 JAL04498.1 GDIQ01071135 GDIQ01060454 JAN34283.1 GDHF01023927 JAI28387.1 GDRN01109716 JAI57038.1 GDIQ01031064 JAN63673.1 GDHF01015858 JAI36456.1 GDIP01149084 JAJ74318.1 GDIQ01058099 JAN36638.1 GDIP01035101 JAM68614.1 GDIQ01031063 JAN63674.1 GDIQ01235889 JAK15836.1 GL732584 EFX74231.1 LRGB01002860 KZS05917.1 GDIQ01018683 JAN76054.1
RVE50892.1 NWSH01003215 PCG66756.1 JTDY01006025 KOB66355.1 KQ459595 KPI95638.1 JTDY01006550 KOB65899.1 KQ461154 KPJ08565.1 GEHC01000166 JAV47479.1 GEDC01007011 JAS30287.1 ATLV01023334 KE525342 KFB48933.1 GECU01011782 JAS95924.1 GEDC01027228 JAS10070.1 AAAB01008963 EAA12089.5 APCN01004856 GEZM01001268 GEZM01001267 JAV98007.1 ADMH02000457 ETN66414.1 KQ434893 KZC10657.1 GFDF01001267 JAV12817.1 GFDF01001279 JAV12805.1 AXCN02002173 KQ414685 KOC63852.1 DS232576 EDS43602.1 GBBI01003926 JAC14786.1 NEVH01009069 PNF33972.1 QOIP01000006 RLU21214.1 GANO01002943 JAB56928.1 KQ981193 KYN45136.1 KQ435840 KOX71346.1 KK107039 EZA61886.1 GL888900 EGI57454.1 GL770322 EFZ10192.1 KZ288232 PBC31469.1 KQ971362 EFA08906.2 NNAY01002686 OXU20775.1 GL436710 EFN71634.1 KQ980900 KYN11703.1 ADTU01026413 ADTU01026414 KQ978501 KYM93599.1 KB632356 ERL93407.1 APGK01041542 APGK01041543 KB740994 ENN75953.1 GL448039 EFN85474.1 KK852817 KDR15749.1 KQ976417 KYM89948.1 KQ760221 OAD61360.1 AJVK01012183 AJVK01012184 AJVK01012185 AJVK01012186 AXCM01003938 GBXI01011492 JAD02800.1 GBXI01015214 JAC99077.1 GBXI01010591 GBXI01009712 GBXI01006454 JAD03701.1 JAD04580.1 JAD07838.1 GBYB01000838 JAG70605.1 GAMC01013620 GAMC01013618 JAB92935.1 KA644642 AFP59271.1 AJWK01032855 KB201890 ESO93724.1 GAMC01013619 JAB92936.1 GAMC01013621 JAB92934.1 GAKP01016266 JAC42686.1 KK854575 PTY17025.1 PNF33971.1 GAKP01016263 JAC42689.1 GDHF01023525 JAI28789.1 GDIQ01092085 JAL59641.1 GDIQ01147228 JAL04498.1 GDIQ01071135 GDIQ01060454 JAN34283.1 GDHF01023927 JAI28387.1 GDRN01109716 JAI57038.1 GDIQ01031064 JAN63673.1 GDHF01015858 JAI36456.1 GDIP01149084 JAJ74318.1 GDIQ01058099 JAN36638.1 GDIP01035101 JAM68614.1 GDIQ01031063 JAN63674.1 GDIQ01235889 JAK15836.1 GL732584 EFX74231.1 LRGB01002860 KZS05917.1 GDIQ01018683 JAN76054.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000218220
UP000037510
UP000053268
+ More
UP000192223 UP000053240 UP000030765 UP000075882 UP000007062 UP000076407 UP000075903 UP000075840 UP000075902 UP000075880 UP000000673 UP000075884 UP000069272 UP000076502 UP000075900 UP000075886 UP000053825 UP000076408 UP000075881 UP000002320 UP000005203 UP000002358 UP000235965 UP000279307 UP000078541 UP000075885 UP000053105 UP000053097 UP000007755 UP000242457 UP000007266 UP000215335 UP000000311 UP000078492 UP000005205 UP000078542 UP000030742 UP000019118 UP000075901 UP000008237 UP000027135 UP000078540 UP000095300 UP000092462 UP000075883 UP000075920 UP000095301 UP000092461 UP000030746 UP000000305 UP000076858
UP000192223 UP000053240 UP000030765 UP000075882 UP000007062 UP000076407 UP000075903 UP000075840 UP000075902 UP000075880 UP000000673 UP000075884 UP000069272 UP000076502 UP000075900 UP000075886 UP000053825 UP000076408 UP000075881 UP000002320 UP000005203 UP000002358 UP000235965 UP000279307 UP000078541 UP000075885 UP000053105 UP000053097 UP000007755 UP000242457 UP000007266 UP000215335 UP000000311 UP000078492 UP000005205 UP000078542 UP000030742 UP000019118 UP000075901 UP000008237 UP000027135 UP000078540 UP000095300 UP000092462 UP000075883 UP000075920 UP000095301 UP000092461 UP000030746 UP000000305 UP000076858
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9J708
A0A2H1VR65
A0A212FL94
A0A3S2M4R0
A0A2A4J4U1
A0A0L7KTC2
+ More
A0A194PRR0 A0A0L7KS97 A0A1W4W5B0 A0A1W4WGT8 A0A194QUB0 A0A1W7R8L0 A0A1B6DXA4 A0A084WFD9 A0A1B6J9Q8 A0A1B6C9Z6 A0A182LLY2 A0A1S4GA40 Q7Q4V8 A0A182WZ83 A0A182V524 A0A182HMQ2 A0A1Y1NPC6 A0A182THA0 A0A182ITF2 W5JUK0 A0A182N129 A0A182FJI4 A0A154PFR4 A0A1L8E287 A0A1L8E2T4 A0A182R242 A0A182QSP5 A0A0L7QZ87 A0A182XYC3 A0A182K915 B0XAA5 A0A023F196 A0A088AQ38 K7IVN9 A0A2J7QZH2 A0A3L8DL94 U5ET38 A0A195FXI2 A0A182PUB7 A0A0M8ZW98 A0A026X1J2 F4X7W8 E9JAF8 A0A2A3EJU3 D6WXU9 A0A232EQX8 E2A4N2 A0A151IVG1 A0A158NUF5 A0A195BY63 U4UJX4 N6TDX5 A0A182T6J5 E2BFT6 A0A067R874 A0A195BTE4 A0A310SUV0 A0A1I8Q012 A0A1B0EW75 A0A182LRX3 A0A1I8Q008 A0A1I8PZY1 A0A0A1WWD9 A0A0A1WKZ4 A0A0A1XBJ8 A0A0C9PIR3 W8AVQ0 A0A182VVH1 T1P7W8 A0A1I8N6G7 A0A1B0CWU0 V4BXA9 W8BI70 W8BUZ8 A0A034VHE3 A0A2R7WF77 A0A2J7QZE7 A0A034VJQ4 A0A0K8UQ34 A0A0P5TFY2 A0A0P5N4Y6 A0A0P6EPA7 A0A0K8UNY1 A0A0P4W215 A0A0P6GP16 A0A0K8VC77 A0A0P5EFG0 A0A0P6ETL0 A0A0P6A4P7 A0A0P6GRV0 A0A0P5GVJ6 E9H1T9 A0A164NFS1 A0A0P6HKY5
A0A194PRR0 A0A0L7KS97 A0A1W4W5B0 A0A1W4WGT8 A0A194QUB0 A0A1W7R8L0 A0A1B6DXA4 A0A084WFD9 A0A1B6J9Q8 A0A1B6C9Z6 A0A182LLY2 A0A1S4GA40 Q7Q4V8 A0A182WZ83 A0A182V524 A0A182HMQ2 A0A1Y1NPC6 A0A182THA0 A0A182ITF2 W5JUK0 A0A182N129 A0A182FJI4 A0A154PFR4 A0A1L8E287 A0A1L8E2T4 A0A182R242 A0A182QSP5 A0A0L7QZ87 A0A182XYC3 A0A182K915 B0XAA5 A0A023F196 A0A088AQ38 K7IVN9 A0A2J7QZH2 A0A3L8DL94 U5ET38 A0A195FXI2 A0A182PUB7 A0A0M8ZW98 A0A026X1J2 F4X7W8 E9JAF8 A0A2A3EJU3 D6WXU9 A0A232EQX8 E2A4N2 A0A151IVG1 A0A158NUF5 A0A195BY63 U4UJX4 N6TDX5 A0A182T6J5 E2BFT6 A0A067R874 A0A195BTE4 A0A310SUV0 A0A1I8Q012 A0A1B0EW75 A0A182LRX3 A0A1I8Q008 A0A1I8PZY1 A0A0A1WWD9 A0A0A1WKZ4 A0A0A1XBJ8 A0A0C9PIR3 W8AVQ0 A0A182VVH1 T1P7W8 A0A1I8N6G7 A0A1B0CWU0 V4BXA9 W8BI70 W8BUZ8 A0A034VHE3 A0A2R7WF77 A0A2J7QZE7 A0A034VJQ4 A0A0K8UQ34 A0A0P5TFY2 A0A0P5N4Y6 A0A0P6EPA7 A0A0K8UNY1 A0A0P4W215 A0A0P6GP16 A0A0K8VC77 A0A0P5EFG0 A0A0P6ETL0 A0A0P6A4P7 A0A0P6GRV0 A0A0P5GVJ6 E9H1T9 A0A164NFS1 A0A0P6HKY5
PDB
5V4Q
E-value=2.89142e-75,
Score=719
Ontologies
PATHWAY
GO
PANTHER
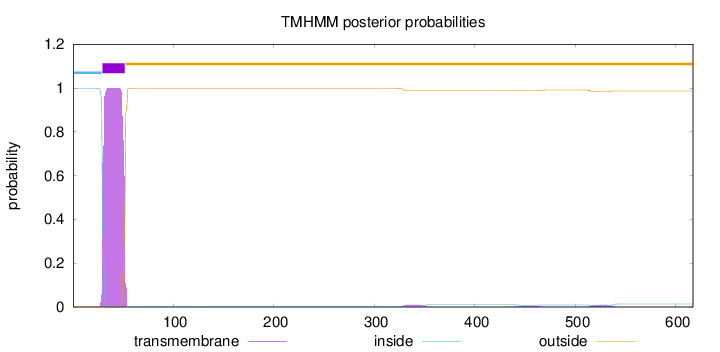
Topology
Length:
617
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.35291
Exp number, first 60 AAs:
21.83053
Total prob of N-in:
0.99875
POSSIBLE N-term signal
sequence
inside
1 - 29
TMhelix
30 - 52
outside
53 - 617
Population Genetic Test Statistics
Pi
266.399497
Theta
183.951312
Tajima's D
1.632535
CLR
0.33771
CSRT
0.812059397030149
Interpretation
Uncertain
