Gene
KWMTBOMO04653
Pre Gene Modal
BGIBMGA005512
Annotation
PREDICTED:_sec1_family_domain-containing_protein_2-like_[Amyelois_transitella]
Full name
Sec1 family domain-containing protein 2
Alternative Name
Neuronal Sec1
Syntaxin-binding protein 1-like 1
Syntaxin-binding protein 1-like 1
Location in the cell
Cytoplasmic Reliability : 1.163 Nuclear Reliability : 1.792
Sequence
CDS
ATGTCCGATTCATCTGTAAAAGAATTCTGTAAGACTTGGTGGGCCGAAGTTTACAACAGAATTATTGGTGCTGTTGTCTTCTTGGATGATGCTAGTTCCGAATGTCTTCATTGGGATGGGGGCCTTTTCAACCTGCTCGAAGGAGGTGCTGTTGCAGTTAAAAGTTTATCTCCATTTGAGTGTGCGACAAAAGATCAAAGGAAAGGAGTATTCATAACCCAGACTACTTCCACACAACTACGTACAATACAAGACATTATCAAACACAGTGATTTCTCACATTGTACACTGATATCATGTGTCAGTTTGGATGTTGTATACTTGGAGCTCCATGAAGGTAAAGATGTGAGTGATGTCATCACTGCTGGAAGAGGGCCTGCGGAGGCTACTAAGATGCTGGAGGATATGATGGTTCAGTGGATCGGAAGAAGTCCATGTGCCGTCCAAGTAGTGCACATACCGATCTTCACAATTTCTCCAACAAATGTAGTATTTCTGACTCCACCATTTCATGCCATGTTCCCTCTCTATGATGGTAGACTGACACCGGAGACAACTTCAGTAGATCTATATACTTTAAAGAGTGAGGAAAGATCTCAGATAAGAAGATTAGCCAGTTGTCTTAATAGCATGTTTGAATCAATGAACTTAAAAGAGGATGTCTATTACATGGGAACCTATAGCTCTTTATTAGCTGGTGTACTAGAAAATTCACCTGTTTGTCTGTCAAGAAGAAAGAATTGCACGAATCCTGCGTCATTAATAATAGTGGACCGAACCTTAGATCTCTGCAGTGTGACAAGTCATTCTATGGAATCTGTTCTGGACAAAGTGTTGGCAGTTTTACCAAAGTTCCCAGGGCATTCGAATGAAGTTGCCGTTGATATGTCTCCGCTTTGTGAAGCTAATGTAATCACCCATCCAGATGACCTACAACTAAGCCCGGGCTGTCTCTACCACGCGAACGATGATATATGCACACAAACCTTTGAACACATGCTAAACAAGTCTCAAAAGGAAGTGATGTTTGATTTATACAATAAATTGTCAAAGTTGGACGTACAGCGGTCACCGAGTCCCAAGACTCTGCTCAGGGTTACTCCACAGAGCGTCGACAAGATCGTTACGGCGGCTAAAGGGAATTATGACGTAATAAGCAATCATCTAGGCATACTACAGCTGGCATTAGCCGTCGTACAAACGTTGAAGTCGCCGAGACGAGCTCAATTGGAACTCCTGGCGAGCTTAGAGAAACAGGTCCTTCAGAATTTGGGTGCCAGCCGAGACTCCACTAGTGTTTTGCAACAGATGAGTACATTAATAAAATCCCGTAAAGAACGCAATTTGCCTTTGGATAATCTGCTCGCGTTAATCATTCACATATACTCTCTCGGCGGTACTGAAGTCACATTCTCAAAAACTCATGAACAGGATCTAATTGAGGCTCTCAGTGTTGCCATGTTCGAAGATCACAAAAGCTTATTTCCTCAAGTTGAAGGGCATATAGTAACACCGGAGGAATGTGATGAGATTTCTCAAAAAACATTGAAGCGACTAAAGGAAATAGCATTGATGAGGAAAAATTTGCAGAAATACAACTCGTTAATAAAGACAAGCGAGACAGGAATCGGGCACGAATACCGCGGCCTGTTACTGCAGCTAGTCGATGACCTCGTGGACACGGATAGACCAGAACTAACAGACCTGCGACACAGAAATGAGGGTATCAAAGATTTGCTGAGAAGCGGCTTGAACATCTTGACAAGCAAGAAATCTCGCACTTTGAAACATCCACTCGAGAGTCCCACTGTGATTTTGTTTGTGATTGGAGGCGTGACCGCTGAGGAATGCAAGCGTCTGCACCGCAGCGTCATCACGAGCGGCGTCGACAACGCCGTGCTCATCGGGGCCACTAAGTTCGTAACGCCGGTCGAAGCTATGAAAGATGTTTTGCTTTTATAG
Protein
MSDSSVKEFCKTWWAEVYNRIIGAVVFLDDASSECLHWDGGLFNLLEGGAVAVKSLSPFECATKDQRKGVFITQTTSTQLRTIQDIIKHSDFSHCTLISCVSLDVVYLELHEGKDVSDVITAGRGPAEATKMLEDMMVQWIGRSPCAVQVVHIPIFTISPTNVVFLTPPFHAMFPLYDGRLTPETTSVDLYTLKSEERSQIRRLASCLNSMFESMNLKEDVYYMGTYSSLLAGVLENSPVCLSRRKNCTNPASLIIVDRTLDLCSVTSHSMESVLDKVLAVLPKFPGHSNEVAVDMSPLCEANVITHPDDLQLSPGCLYHANDDICTQTFEHMLNKSQKEVMFDLYNKLSKLDVQRSPSPKTLLRVTPQSVDKIVTAAKGNYDVISNHLGILQLALAVVQTLKSPRRAQLELLASLEKQVLQNLGASRDSTSVLQQMSTLIKSRKERNLPLDNLLALIIHIYSLGGTEVTFSKTHEQDLIEALSVAMFEDHKSLFPQVEGHIVTPEECDEISQKTLKRLKEIALMRKNLQKYNSLIKTSETGIGHEYRGLLLQLVDDLVDTDRPELTDLRHRNEGIKDLLRSGLNILTSKKSRTLKHPLESPTVILFVIGGVTAEECKRLHRSVITSGVDNAVLIGATKFVTPVEAMKDVLLL
Summary
Description
May be involved in protein transport.
Similarity
Belongs to the STXBP/unc-18/SEC1 family.
Keywords
Alternative splicing
Complete proteome
Protein transport
Reference proteome
Transport
Feature
chain Sec1 family domain-containing protein 2
splice variant In isoform 4.
splice variant In isoform 4.
Uniprot
H9J7M0
A0A2A4J3N2
A0A194PQP6
A0A194QTC7
A0A212FL99
A0A2H1V2I0
+ More
A0A0L7L2T5 A0A2J7RM52 A0A067R6N7 A0A2J7RM27 A0A0V0G6H3 A0A069DZE7 A0A023F969 A0A0P4VMU9 R4G813 T1J9Z1 H3AEC3 A0A3L8DDA9 A0A026WRT2 A0A151JMS8 F4WKT2 A0A151PAK4 E2AJ45 A0A158P3Z1 A0A151X0H9 A0A195CN69 E2BUX0 A0A151JVL9 A0A1S3JPG4 A0A0L8GSS7 I3J4K3 A0A1S3J8I2 A0A1S3J913 A0A3B4FW51 A0A3P8Q314 A0A232FD87 K9IMB6 E9J1R1 A0A2G8JSZ3 A0A3P9CZC2 A0A3B3CB14 F1RBN1 A0A384A0G9 A0A3S2PZ18 K7IMU8 A0A2Y9DXK4 A0A1S3PA72 A0A210R165 A0JN38 Q5U2R9 A0A384A0I3 A0A2C9KS83 A0A1L8HTQ6 A0A2Y9MLY1 A0A3Q3G8Z2 H2UMU3 W4Y112 A0A1A7WXU0 A0A3Q3M4J4 A0A310SP04 A0A340X3K5 V3Z5D0 A0A1U7QTX2 A0A1S3KIM4 A0A154PNC5 A0A195BKZ3 W5N9U0 G1SDX5 A0A1A8FF21 A0A341BL70 A0A0P5S609 A0A1A8IE02 A0A0P5QIG0 A0A0P5S4C2 A0A2Y9MCU1 W5QBH7 A0A2I4CS52 A0A2U3VMP3 A0A1S3EK51 A0A0J7KKB0 A0A0N7ZIZ4 Q8BTY8 A0A0P6HN87 A0A1A8PV10 L7MI57 A0A384CXX1 A0A3Q1HYH5 A0A2Y9JL20 A0A3Q0G381 Q3UPD0 A0A2Y9MGA5 A0A0P4VYK3 A0A2Y9I062 A0A2U3XSL0 A0A224Z8D1 A0A286XJX5 A0A0P5SNS9 A0A3B3RH14 A0A1A8QV22 A0A3Q7WI06
A0A0L7L2T5 A0A2J7RM52 A0A067R6N7 A0A2J7RM27 A0A0V0G6H3 A0A069DZE7 A0A023F969 A0A0P4VMU9 R4G813 T1J9Z1 H3AEC3 A0A3L8DDA9 A0A026WRT2 A0A151JMS8 F4WKT2 A0A151PAK4 E2AJ45 A0A158P3Z1 A0A151X0H9 A0A195CN69 E2BUX0 A0A151JVL9 A0A1S3JPG4 A0A0L8GSS7 I3J4K3 A0A1S3J8I2 A0A1S3J913 A0A3B4FW51 A0A3P8Q314 A0A232FD87 K9IMB6 E9J1R1 A0A2G8JSZ3 A0A3P9CZC2 A0A3B3CB14 F1RBN1 A0A384A0G9 A0A3S2PZ18 K7IMU8 A0A2Y9DXK4 A0A1S3PA72 A0A210R165 A0JN38 Q5U2R9 A0A384A0I3 A0A2C9KS83 A0A1L8HTQ6 A0A2Y9MLY1 A0A3Q3G8Z2 H2UMU3 W4Y112 A0A1A7WXU0 A0A3Q3M4J4 A0A310SP04 A0A340X3K5 V3Z5D0 A0A1U7QTX2 A0A1S3KIM4 A0A154PNC5 A0A195BKZ3 W5N9U0 G1SDX5 A0A1A8FF21 A0A341BL70 A0A0P5S609 A0A1A8IE02 A0A0P5QIG0 A0A0P5S4C2 A0A2Y9MCU1 W5QBH7 A0A2I4CS52 A0A2U3VMP3 A0A1S3EK51 A0A0J7KKB0 A0A0N7ZIZ4 Q8BTY8 A0A0P6HN87 A0A1A8PV10 L7MI57 A0A384CXX1 A0A3Q1HYH5 A0A2Y9JL20 A0A3Q0G381 Q3UPD0 A0A2Y9MGA5 A0A0P4VYK3 A0A2Y9I062 A0A2U3XSL0 A0A224Z8D1 A0A286XJX5 A0A0P5SNS9 A0A3B3RH14 A0A1A8QV22 A0A3Q7WI06
Pubmed
19121390
26354079
22118469
26227816
24845553
26334808
+ More
25474469 27129103 9215903 30249741 24508170 21719571 22293439 20798317 21347285 25186727 26383154 28648823 21282665 29023486 29451363 23594743 20075255 28812685 19393038 15489334 15057822 15562597 27762356 21551351 23254933 21993624 20809919 16141072 21183079 25576852 10349636 11042159 11076861 11217851 12466851 16141073 28797301 29240929
25474469 27129103 9215903 30249741 24508170 21719571 22293439 20798317 21347285 25186727 26383154 28648823 21282665 29023486 29451363 23594743 20075255 28812685 19393038 15489334 15057822 15562597 27762356 21551351 23254933 21993624 20809919 16141072 21183079 25576852 10349636 11042159 11076861 11217851 12466851 16141073 28797301 29240929
EMBL
BABH01014021
NWSH01003215
PCG66757.1
KQ459595
KPI95637.1
KQ461154
+ More
KPJ08564.1 AGBW02007817 OWR54512.1 ODYU01000166 SOQ34474.1 JTDY01003415 KOB69594.1 NEVH01002577 PNF41889.1 KK852705 KDR18013.1 PNF41887.1 GECL01002502 JAP03622.1 GBGD01000770 JAC88119.1 GBBI01001259 JAC17453.1 GDKW01000706 JAI55889.1 GAHY01001772 JAA75738.1 JH431980 AFYH01128740 AFYH01128741 AFYH01128742 AFYH01128743 AFYH01128744 AFYH01128745 AFYH01128746 AFYH01128747 AFYH01128748 AFYH01128749 QOIP01000009 RLU18374.1 KK107119 EZA58732.1 KQ978949 KYN27363.1 GL888206 EGI65174.1 AKHW03000533 KYO46000.1 GL439967 EFN66485.1 ADTU01008623 KQ982612 KYQ53880.1 KQ977513 KYN02158.1 GL450772 EFN80507.1 KQ981701 KYN37504.1 KQ420672 KOF79650.1 AERX01004391 AERX01004392 AERX01004393 AERX01004394 AERX01004395 AERX01004396 NNAY01000428 OXU28463.1 GABZ01004797 JAA48728.1 GL767674 EFZ13181.1 MRZV01001308 PIK38839.1 BX511028 CABZ01039446 CABZ01039447 CABZ01039448 CABZ01079987 CU693368 LO018613 CM012440 RVE73817.1 NEDP02000942 OWF54641.1 BC126501 AAI26502.1 AABR07014890 AABR07014891 AABR07072395 AABR07072396 AC099101 AC114452 AC128952 BC085889 AAH85889.1 CM004466 OCT99496.1 AAGJ04127454 HADW01009118 HADX01011269 SBP10518.1 KQ761783 OAD57062.1 KB203219 ESO85938.1 KQ434998 KZC13343.1 KQ976453 KYM85352.1 AHAT01002256 AHAT01002257 AHAT01002258 AHAT01002259 AHAT01002260 AHAT01002261 AHAT01002262 HAEB01011100 HAEC01002655 SBQ57627.1 GDIQ01109036 JAL42690.1 HAED01008927 HAEE01017079 SBQ95139.1 GDIQ01120133 JAL31593.1 GDIQ01092590 JAL59136.1 AMGL01098545 AMGL01098546 AMGL01098547 AMGL01098548 AMGL01098549 AMGL01098550 AMGL01098551 AMGL01098552 AMGL01098553 AMGL01098554 LBMM01006176 KMQ90853.1 GDIP01240785 JAI82616.1 AK036637 AK044408 AK047026 AK047738 AK050673 AK054548 AK088353 AK089011 AK140246 BC042528 GDIQ01024653 JAN70084.1 HAEF01015428 HAEG01009570 SBR84852.1 GACK01001532 JAA63502.1 AK143615 BAE25466.1 GDRN01087114 JAI61076.1 GFPF01011604 MAA22750.1 AAKN02028258 AAKN02028259 AAKN02028260 AAKN02028261 AAKN02028262 AAKN02028263 GDIQ01101593 JAL50133.1 HAEH01013324 HAEI01016237 SBR97108.1
KPJ08564.1 AGBW02007817 OWR54512.1 ODYU01000166 SOQ34474.1 JTDY01003415 KOB69594.1 NEVH01002577 PNF41889.1 KK852705 KDR18013.1 PNF41887.1 GECL01002502 JAP03622.1 GBGD01000770 JAC88119.1 GBBI01001259 JAC17453.1 GDKW01000706 JAI55889.1 GAHY01001772 JAA75738.1 JH431980 AFYH01128740 AFYH01128741 AFYH01128742 AFYH01128743 AFYH01128744 AFYH01128745 AFYH01128746 AFYH01128747 AFYH01128748 AFYH01128749 QOIP01000009 RLU18374.1 KK107119 EZA58732.1 KQ978949 KYN27363.1 GL888206 EGI65174.1 AKHW03000533 KYO46000.1 GL439967 EFN66485.1 ADTU01008623 KQ982612 KYQ53880.1 KQ977513 KYN02158.1 GL450772 EFN80507.1 KQ981701 KYN37504.1 KQ420672 KOF79650.1 AERX01004391 AERX01004392 AERX01004393 AERX01004394 AERX01004395 AERX01004396 NNAY01000428 OXU28463.1 GABZ01004797 JAA48728.1 GL767674 EFZ13181.1 MRZV01001308 PIK38839.1 BX511028 CABZ01039446 CABZ01039447 CABZ01039448 CABZ01079987 CU693368 LO018613 CM012440 RVE73817.1 NEDP02000942 OWF54641.1 BC126501 AAI26502.1 AABR07014890 AABR07014891 AABR07072395 AABR07072396 AC099101 AC114452 AC128952 BC085889 AAH85889.1 CM004466 OCT99496.1 AAGJ04127454 HADW01009118 HADX01011269 SBP10518.1 KQ761783 OAD57062.1 KB203219 ESO85938.1 KQ434998 KZC13343.1 KQ976453 KYM85352.1 AHAT01002256 AHAT01002257 AHAT01002258 AHAT01002259 AHAT01002260 AHAT01002261 AHAT01002262 HAEB01011100 HAEC01002655 SBQ57627.1 GDIQ01109036 JAL42690.1 HAED01008927 HAEE01017079 SBQ95139.1 GDIQ01120133 JAL31593.1 GDIQ01092590 JAL59136.1 AMGL01098545 AMGL01098546 AMGL01098547 AMGL01098548 AMGL01098549 AMGL01098550 AMGL01098551 AMGL01098552 AMGL01098553 AMGL01098554 LBMM01006176 KMQ90853.1 GDIP01240785 JAI82616.1 AK036637 AK044408 AK047026 AK047738 AK050673 AK054548 AK088353 AK089011 AK140246 BC042528 GDIQ01024653 JAN70084.1 HAEF01015428 HAEG01009570 SBR84852.1 GACK01001532 JAA63502.1 AK143615 BAE25466.1 GDRN01087114 JAI61076.1 GFPF01011604 MAA22750.1 AAKN02028258 AAKN02028259 AAKN02028260 AAKN02028261 AAKN02028262 AAKN02028263 GDIQ01101593 JAL50133.1 HAEH01013324 HAEI01016237 SBR97108.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000235965 UP000027135 UP000008672 UP000279307 UP000053097 UP000078492 UP000007755 UP000050525 UP000000311 UP000005205 UP000075809 UP000078542 UP000008237 UP000078541 UP000085678 UP000053454 UP000005207 UP000261460 UP000265100 UP000215335 UP000230750 UP000265160 UP000261560 UP000000437 UP000261681 UP000002358 UP000248480 UP000087266 UP000242188 UP000009136 UP000002494 UP000076420 UP000186698 UP000248483 UP000261660 UP000005226 UP000007110 UP000261640 UP000265300 UP000030746 UP000189706 UP000076502 UP000078540 UP000018468 UP000001811 UP000252040 UP000002356 UP000192220 UP000245340 UP000081671 UP000036403 UP000000589 UP000261680 UP000257200 UP000248482 UP000189705 UP000248481 UP000245341 UP000005447 UP000261540 UP000286642
UP000235965 UP000027135 UP000008672 UP000279307 UP000053097 UP000078492 UP000007755 UP000050525 UP000000311 UP000005205 UP000075809 UP000078542 UP000008237 UP000078541 UP000085678 UP000053454 UP000005207 UP000261460 UP000265100 UP000215335 UP000230750 UP000265160 UP000261560 UP000000437 UP000261681 UP000002358 UP000248480 UP000087266 UP000242188 UP000009136 UP000002494 UP000076420 UP000186698 UP000248483 UP000261660 UP000005226 UP000007110 UP000261640 UP000265300 UP000030746 UP000189706 UP000076502 UP000078540 UP000018468 UP000001811 UP000252040 UP000002356 UP000192220 UP000245340 UP000081671 UP000036403 UP000000589 UP000261680 UP000257200 UP000248482 UP000189705 UP000248481 UP000245341 UP000005447 UP000261540 UP000286642
Pfam
PF00995 Sec1
SUPFAM
SSF56815
SSF56815
Gene 3D
ProteinModelPortal
H9J7M0
A0A2A4J3N2
A0A194PQP6
A0A194QTC7
A0A212FL99
A0A2H1V2I0
+ More
A0A0L7L2T5 A0A2J7RM52 A0A067R6N7 A0A2J7RM27 A0A0V0G6H3 A0A069DZE7 A0A023F969 A0A0P4VMU9 R4G813 T1J9Z1 H3AEC3 A0A3L8DDA9 A0A026WRT2 A0A151JMS8 F4WKT2 A0A151PAK4 E2AJ45 A0A158P3Z1 A0A151X0H9 A0A195CN69 E2BUX0 A0A151JVL9 A0A1S3JPG4 A0A0L8GSS7 I3J4K3 A0A1S3J8I2 A0A1S3J913 A0A3B4FW51 A0A3P8Q314 A0A232FD87 K9IMB6 E9J1R1 A0A2G8JSZ3 A0A3P9CZC2 A0A3B3CB14 F1RBN1 A0A384A0G9 A0A3S2PZ18 K7IMU8 A0A2Y9DXK4 A0A1S3PA72 A0A210R165 A0JN38 Q5U2R9 A0A384A0I3 A0A2C9KS83 A0A1L8HTQ6 A0A2Y9MLY1 A0A3Q3G8Z2 H2UMU3 W4Y112 A0A1A7WXU0 A0A3Q3M4J4 A0A310SP04 A0A340X3K5 V3Z5D0 A0A1U7QTX2 A0A1S3KIM4 A0A154PNC5 A0A195BKZ3 W5N9U0 G1SDX5 A0A1A8FF21 A0A341BL70 A0A0P5S609 A0A1A8IE02 A0A0P5QIG0 A0A0P5S4C2 A0A2Y9MCU1 W5QBH7 A0A2I4CS52 A0A2U3VMP3 A0A1S3EK51 A0A0J7KKB0 A0A0N7ZIZ4 Q8BTY8 A0A0P6HN87 A0A1A8PV10 L7MI57 A0A384CXX1 A0A3Q1HYH5 A0A2Y9JL20 A0A3Q0G381 Q3UPD0 A0A2Y9MGA5 A0A0P4VYK3 A0A2Y9I062 A0A2U3XSL0 A0A224Z8D1 A0A286XJX5 A0A0P5SNS9 A0A3B3RH14 A0A1A8QV22 A0A3Q7WI06
A0A0L7L2T5 A0A2J7RM52 A0A067R6N7 A0A2J7RM27 A0A0V0G6H3 A0A069DZE7 A0A023F969 A0A0P4VMU9 R4G813 T1J9Z1 H3AEC3 A0A3L8DDA9 A0A026WRT2 A0A151JMS8 F4WKT2 A0A151PAK4 E2AJ45 A0A158P3Z1 A0A151X0H9 A0A195CN69 E2BUX0 A0A151JVL9 A0A1S3JPG4 A0A0L8GSS7 I3J4K3 A0A1S3J8I2 A0A1S3J913 A0A3B4FW51 A0A3P8Q314 A0A232FD87 K9IMB6 E9J1R1 A0A2G8JSZ3 A0A3P9CZC2 A0A3B3CB14 F1RBN1 A0A384A0G9 A0A3S2PZ18 K7IMU8 A0A2Y9DXK4 A0A1S3PA72 A0A210R165 A0JN38 Q5U2R9 A0A384A0I3 A0A2C9KS83 A0A1L8HTQ6 A0A2Y9MLY1 A0A3Q3G8Z2 H2UMU3 W4Y112 A0A1A7WXU0 A0A3Q3M4J4 A0A310SP04 A0A340X3K5 V3Z5D0 A0A1U7QTX2 A0A1S3KIM4 A0A154PNC5 A0A195BKZ3 W5N9U0 G1SDX5 A0A1A8FF21 A0A341BL70 A0A0P5S609 A0A1A8IE02 A0A0P5QIG0 A0A0P5S4C2 A0A2Y9MCU1 W5QBH7 A0A2I4CS52 A0A2U3VMP3 A0A1S3EK51 A0A0J7KKB0 A0A0N7ZIZ4 Q8BTY8 A0A0P6HN87 A0A1A8PV10 L7MI57 A0A384CXX1 A0A3Q1HYH5 A0A2Y9JL20 A0A3Q0G381 Q3UPD0 A0A2Y9MGA5 A0A0P4VYK3 A0A2Y9I062 A0A2U3XSL0 A0A224Z8D1 A0A286XJX5 A0A0P5SNS9 A0A3B3RH14 A0A1A8QV22 A0A3Q7WI06
Ontologies
GO
PANTHER
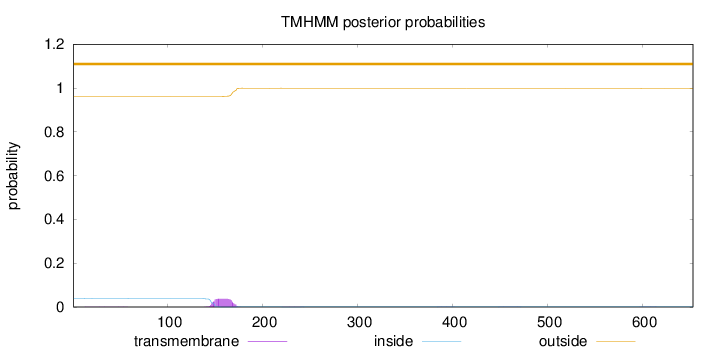
Topology
Length:
653
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.889300000000001
Exp number, first 60 AAs:
0.01209
Total prob of N-in:
0.03858
outside
1 - 653
Population Genetic Test Statistics
Pi
220.357943
Theta
164.509203
Tajima's D
1.377622
CLR
0.272198
CSRT
0.754562271886406
Interpretation
Uncertain
