Pre Gene Modal
BGIBMGA005508
Annotation
PREDICTED:_bifunctional_coenzyme_A_synthase_isoform_X1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.781 Mitochondrial Reliability : 1.624
Sequence
CDS
ATGGCACATAATGGCCTCTTGTTTGTATCAAATGCTGCAAAAGCACACATTGCCTGTATGAAGGCAGCTAAGTATGTGCGGAATGTTATGTATGTGAAGATTAAAATAAATGCTGAGAATTCTTCACAAGTGATCGGTAGACAAATTGTAAACATTTATACAAAGGCTCAAACCAATATAGACATAAGGTTAATGGTGAAACCATATGAAAAAAGTAAGGATATTATCACACGTCAACCAATTGATTTGATTATGTATGACAGTGAATCAGCTAAAGAAGTCGAAGTTTTGAGAAAGTCAGTGAAGTCTCTGGACAGAGGATACAGATTGCAGAATATAGACACCAGTGACTCACCGATAGAGAATAATGTCAATGAAGAAGTAAAGGCGTATGAGTATGTTGCTCTGGGAGGGACATTTGACCGACTGCATAATGGACACAAGATATTGCTGTCACAAGCTGTGCTGCGCTCCACTAGGCATGTCACGGTGGGCGTCACTGATGTTAATATGATTCAGTCCAAGAAGCTATGGGAATTGATTGAACCAGTTGAAAAAAGGATTGAAACTGTTCTACAATTTCTAAACGATATAAATCCAGACTTGGAATACAATGTTTTTCCGCTGAAAGATCTTTATGGACCTACAAAAGATGATCCCAAATTTCAGTTAATAGTTGTTAGCGAAGAGACCGTCCGAGGTGCAGTGAAAATCAATGAAAAACGTCAAGAGAAAAACTTAAAAAAATTAGACACGTACGTCATAGAATTGGCGCAGGACTCGCATCCGAGAAGACATCACGAGGAAGAAGACAAAGTCAGCTCGAGTAATCAGAGAATGAGATTGTTGGGTACTCTACTCAGACCTCCCAGGCCCAATCCAAATATACCGGATTGGCCGTACGTGATCGGTTTGGCGGGCGGCATCGCCAGCGGCAAGAGTAATATTGCGGAAAAACTCAAATCAAAAGGTGCGGCGATAATAAACTGTGATATAATAGCCCACGAGCTCTACAGGCCGGGGTTGCCGCTCAACACGACTATAGCGCAAAGCTTCGGTGACCACGTCATCACATCCGACGGCGAAGTGGACAGGAGGAAGCTGGGTCAGATTGTTTTTAGTGATAAGAGTCAGCTACAAAAGTTGAATGAACTAGTATGGCCGGCAGTAATCGAAGAGACGCAGAGGAGAGTCGTGGCTCTCGGCAAACAAGGATATAAGGTGGTGGTGATGGAAGCGGCTGTGATGGTGCGAGCTAAGTGGTACGAAAGATGTCATCAGCTATGGGCGGTGATCATACCGCCGGATGAGGCCATAAAAAGGCTGCAAAAACGCAATGGACTGACCGAAGAAGAAGCGAGACAGCGTATAGAGGCCCAACCCTCGAATTCGGAACAAGTGGCTCACGCCAATATAGTGTTCAGTCCGTTCTGGAGCTACGAATATACGCAATCGCAAATCGATAGAGCATGGCAACAATTAAATGAATACCTGGACAACAAGAAGTGA
Protein
MAHNGLLFVSNAAKAHIACMKAAKYVRNVMYVKIKINAENSSQVIGRQIVNIYTKAQTNIDIRLMVKPYEKSKDIITRQPIDLIMYDSESAKEVEVLRKSVKSLDRGYRLQNIDTSDSPIENNVNEEVKAYEYVALGGTFDRLHNGHKILLSQAVLRSTRHVTVGVTDVNMIQSKKLWELIEPVEKRIETVLQFLNDINPDLEYNVFPLKDLYGPTKDDPKFQLIVVSEETVRGAVKINEKRQEKNLKKLDTYVIELAQDSHPRRHHEEEDKVSSSNQRMRLLGTLLRPPRPNPNIPDWPYVIGLAGGIASGKSNIAEKLKSKGAAIINCDIIAHELYRPGLPLNTTIAQSFGDHVITSDGEVDRRKLGQIVFSDKSQLQKLNELVWPAVIEETQRRVVALGKQGYKVVVMEAAVMVRAKWYERCHQLWAVIIPPDEAIKRLQKRNGLTEEEARQRIEAQPSNSEQVAHANIVFSPFWSYEYTQSQIDRAWQQLNEYLDNKK
Summary
Uniprot
H9J7L6
A0A3S2NLQ9
A0A194PQA9
A0A194QSR4
A0A212FDA4
A0A2H1VM74
+ More
A0A2A4IWD7 A0A2A4IY13 A0A2A4IXZ9 A0A2A4IXA9 A0A026VX34 E2AXA5 A0A1B6MQ73 E2BI82 A0A1B6JTY9 A0A224XLZ6 A0A069DV06 A0A154PJT0 A0A0P4VZC0 K7J9U3 A0A0J7KN29 A0A0L7QNF6 B4HAC6 A0A067RJM7 E9IEC9 Q2LZB0 A0A195EZR2 A0A1B6CV28 A0A0M8ZSK0 A0A3B0JSH9 F4W8X3 A0A0M4EKG4 A0A158NP16 D6WVB0 A0A195BZG9 A0A2J7R740 A0A195DWU7 A0A2R7WCV1 V5G6G8 B4HUI4 A0A0J9RPD7 A0A2H8TG73 B3M3M7 B4MLR7 U5EW21 J9JKZ4 A0A1L8DZI0 A0A1Y1K7F6 A0A1B0DBI0 Q9VRP4 Q29QT3 A0A1J1IJS8 A0A2J7R728 B4PIW7 B4L992 B0WPT1 T1HTP5 B4LBS1 A0A151X8D2 A0A2A3EKI5 A0A0K8UNB4 A0A1L8DZI6 B4J1J1 A0A2S2QFP9 A0A1W4UX39 J3JW50 B3NC85 A0A1Q3FCS7 A0A0L0CKV1 A0A034WGW8 A0A1B0CL29 A0A182K709 A0A182WK79 A0A182RQ97 A0A1I8P5V1 A0A0K8V5G1 Q0IFZ2 A0A182PN98 N6SZ54 U4US73 W8BVL2 A0A182LJP9 E0V974 Q1HRE4 A0A182LVZ9 A0A182HM55 A0A182TDR7 A0A1I8MTH9 A0A182XCS9 A0A1B6GVX8 A0A182VB58 A0A293M6R1 A0A182GQK9 A0A084W2U6 A0A1B0AQC7 A0A1A9XIM1 Q7Q774 A0A2R5L8N1 A0A182J233 A0A182Y0U4
A0A2A4IWD7 A0A2A4IY13 A0A2A4IXZ9 A0A2A4IXA9 A0A026VX34 E2AXA5 A0A1B6MQ73 E2BI82 A0A1B6JTY9 A0A224XLZ6 A0A069DV06 A0A154PJT0 A0A0P4VZC0 K7J9U3 A0A0J7KN29 A0A0L7QNF6 B4HAC6 A0A067RJM7 E9IEC9 Q2LZB0 A0A195EZR2 A0A1B6CV28 A0A0M8ZSK0 A0A3B0JSH9 F4W8X3 A0A0M4EKG4 A0A158NP16 D6WVB0 A0A195BZG9 A0A2J7R740 A0A195DWU7 A0A2R7WCV1 V5G6G8 B4HUI4 A0A0J9RPD7 A0A2H8TG73 B3M3M7 B4MLR7 U5EW21 J9JKZ4 A0A1L8DZI0 A0A1Y1K7F6 A0A1B0DBI0 Q9VRP4 Q29QT3 A0A1J1IJS8 A0A2J7R728 B4PIW7 B4L992 B0WPT1 T1HTP5 B4LBS1 A0A151X8D2 A0A2A3EKI5 A0A0K8UNB4 A0A1L8DZI6 B4J1J1 A0A2S2QFP9 A0A1W4UX39 J3JW50 B3NC85 A0A1Q3FCS7 A0A0L0CKV1 A0A034WGW8 A0A1B0CL29 A0A182K709 A0A182WK79 A0A182RQ97 A0A1I8P5V1 A0A0K8V5G1 Q0IFZ2 A0A182PN98 N6SZ54 U4US73 W8BVL2 A0A182LJP9 E0V974 Q1HRE4 A0A182LVZ9 A0A182HM55 A0A182TDR7 A0A1I8MTH9 A0A182XCS9 A0A1B6GVX8 A0A182VB58 A0A293M6R1 A0A182GQK9 A0A084W2U6 A0A1B0AQC7 A0A1A9XIM1 Q7Q774 A0A2R5L8N1 A0A182J233 A0A182Y0U4
Pubmed
19121390
26354079
22118469
24508170
30249741
20798317
+ More
26334808 27129103 20075255 17994087 24845553 21282665 15632085 21719571 21347285 18362917 19820115 22936249 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 22516182 26108605 25348373 17510324 23537049 24495485 20966253 20566863 17204158 25315136 26483478 24438588 12364791 14747013 17210077 25244985
26334808 27129103 20075255 17994087 24845553 21282665 15632085 21719571 21347285 18362917 19820115 22936249 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 22516182 26108605 25348373 17510324 23537049 24495485 20966253 20566863 17204158 25315136 26483478 24438588 12364791 14747013 17210077 25244985
EMBL
BABH01014017
RSAL01000041
RVE50887.1
KQ459595
KPI95631.1
KQ461154
+ More
KPJ08558.1 AGBW02009085 OWR51712.1 ODYU01003323 SOQ41923.1 NWSH01005690 PCG64051.1 PCG64050.1 PCG64052.1 PCG64048.1 KK107648 QOIP01000008 EZA48353.1 RLU19780.1 GL443548 EFN61885.1 GEBQ01001938 JAT38039.1 GL448457 EFN84613.1 GECU01005333 JAT02374.1 GFTR01006966 JAW09460.1 GBGD01001124 JAC87765.1 KQ434938 KZC12119.1 GDKW01000663 JAI55932.1 AAZX01007750 AAZX01007849 AAZX01009411 LBMM01005134 KMQ91793.1 KQ414855 KOC60160.1 CH479240 EDW37524.1 KK852470 KDR23193.1 GL762576 EFZ21073.1 CH379069 EAL29598.2 KQ981891 KYN33795.1 GEDC01020031 JAS17267.1 KQ435867 KOX70265.1 OUUW01000009 SPP85074.1 GL887974 EGI69332.1 CP012525 ALC43735.1 ADTU01022032 KQ971363 EFA07751.2 KQ976394 KYM93321.1 NEVH01006738 PNF36642.1 KQ980167 KYN17363.1 KK854635 PTY17502.1 GALX01002791 JAB65675.1 CH480817 EDW50605.1 CM002912 KMY97786.1 GFXV01001246 MBW13051.1 CH902618 EDV40320.1 CH963847 EDW72993.1 GANO01003156 JAB56715.1 ABLF02034945 GFDF01002204 JAV11880.1 GEZM01092875 JAV56431.1 AJVK01013664 AE014296 AAF50749.2 BT024307 ABC86369.1 CVRI01000053 CRK99796.1 PNF36641.1 CM000159 EDW93537.1 CH933816 EDW17267.1 DS232029 EDS32496.1 ACPB03014260 CH940647 EDW68698.1 KQ982422 KYQ56622.1 KZ288223 PBC32004.1 GDHF01024142 JAI28172.1 GFDF01002203 JAV11881.1 CH916366 EDV95882.1 GGMS01007353 MBY76556.1 BT127468 AEE62430.1 CH954178 EDV51043.1 GFDL01009669 JAV25376.1 JRES01000341 KNC32104.1 GAKP01004146 JAC54806.1 AJWK01016860 AJWK01016861 AJWK01016862 GDHF01018326 JAI33988.1 CH477281 EAT44932.1 APGK01058607 KB741291 ENN70523.1 KB632343 ERL93021.1 GAMC01003313 JAC03243.1 DS234989 EEB09930.1 DQ440150 ABF18183.1 AXCM01001822 APCN01004355 GECZ01003266 JAS66503.1 GFWV01010071 MAA34800.1 JXUM01015651 KQ560436 KXJ82428.1 ATLV01019698 KE525278 KFB44540.1 JXJN01001810 AAAB01008960 EAA11969.3 GGLE01001704 MBY05830.1
KPJ08558.1 AGBW02009085 OWR51712.1 ODYU01003323 SOQ41923.1 NWSH01005690 PCG64051.1 PCG64050.1 PCG64052.1 PCG64048.1 KK107648 QOIP01000008 EZA48353.1 RLU19780.1 GL443548 EFN61885.1 GEBQ01001938 JAT38039.1 GL448457 EFN84613.1 GECU01005333 JAT02374.1 GFTR01006966 JAW09460.1 GBGD01001124 JAC87765.1 KQ434938 KZC12119.1 GDKW01000663 JAI55932.1 AAZX01007750 AAZX01007849 AAZX01009411 LBMM01005134 KMQ91793.1 KQ414855 KOC60160.1 CH479240 EDW37524.1 KK852470 KDR23193.1 GL762576 EFZ21073.1 CH379069 EAL29598.2 KQ981891 KYN33795.1 GEDC01020031 JAS17267.1 KQ435867 KOX70265.1 OUUW01000009 SPP85074.1 GL887974 EGI69332.1 CP012525 ALC43735.1 ADTU01022032 KQ971363 EFA07751.2 KQ976394 KYM93321.1 NEVH01006738 PNF36642.1 KQ980167 KYN17363.1 KK854635 PTY17502.1 GALX01002791 JAB65675.1 CH480817 EDW50605.1 CM002912 KMY97786.1 GFXV01001246 MBW13051.1 CH902618 EDV40320.1 CH963847 EDW72993.1 GANO01003156 JAB56715.1 ABLF02034945 GFDF01002204 JAV11880.1 GEZM01092875 JAV56431.1 AJVK01013664 AE014296 AAF50749.2 BT024307 ABC86369.1 CVRI01000053 CRK99796.1 PNF36641.1 CM000159 EDW93537.1 CH933816 EDW17267.1 DS232029 EDS32496.1 ACPB03014260 CH940647 EDW68698.1 KQ982422 KYQ56622.1 KZ288223 PBC32004.1 GDHF01024142 JAI28172.1 GFDF01002203 JAV11881.1 CH916366 EDV95882.1 GGMS01007353 MBY76556.1 BT127468 AEE62430.1 CH954178 EDV51043.1 GFDL01009669 JAV25376.1 JRES01000341 KNC32104.1 GAKP01004146 JAC54806.1 AJWK01016860 AJWK01016861 AJWK01016862 GDHF01018326 JAI33988.1 CH477281 EAT44932.1 APGK01058607 KB741291 ENN70523.1 KB632343 ERL93021.1 GAMC01003313 JAC03243.1 DS234989 EEB09930.1 DQ440150 ABF18183.1 AXCM01001822 APCN01004355 GECZ01003266 JAS66503.1 GFWV01010071 MAA34800.1 JXUM01015651 KQ560436 KXJ82428.1 ATLV01019698 KE525278 KFB44540.1 JXJN01001810 AAAB01008960 EAA11969.3 GGLE01001704 MBY05830.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000007151
UP000218220
+ More
UP000053097 UP000279307 UP000000311 UP000008237 UP000076502 UP000002358 UP000036403 UP000053825 UP000008744 UP000027135 UP000001819 UP000078541 UP000053105 UP000268350 UP000007755 UP000092553 UP000005205 UP000007266 UP000078540 UP000235965 UP000078492 UP000001292 UP000007801 UP000007798 UP000007819 UP000092462 UP000000803 UP000183832 UP000002282 UP000009192 UP000002320 UP000015103 UP000008792 UP000075809 UP000242457 UP000001070 UP000192221 UP000008711 UP000037069 UP000092461 UP000075881 UP000075920 UP000075900 UP000095300 UP000008820 UP000075885 UP000019118 UP000030742 UP000075882 UP000009046 UP000075883 UP000075840 UP000075902 UP000095301 UP000076407 UP000075903 UP000069940 UP000249989 UP000030765 UP000092460 UP000092443 UP000007062 UP000075880 UP000076408
UP000053097 UP000279307 UP000000311 UP000008237 UP000076502 UP000002358 UP000036403 UP000053825 UP000008744 UP000027135 UP000001819 UP000078541 UP000053105 UP000268350 UP000007755 UP000092553 UP000005205 UP000007266 UP000078540 UP000235965 UP000078492 UP000001292 UP000007801 UP000007798 UP000007819 UP000092462 UP000000803 UP000183832 UP000002282 UP000009192 UP000002320 UP000015103 UP000008792 UP000075809 UP000242457 UP000001070 UP000192221 UP000008711 UP000037069 UP000092461 UP000075881 UP000075920 UP000075900 UP000095300 UP000008820 UP000075885 UP000019118 UP000030742 UP000075882 UP000009046 UP000075883 UP000075840 UP000075902 UP000095301 UP000076407 UP000075903 UP000069940 UP000249989 UP000030765 UP000092460 UP000092443 UP000007062 UP000075880 UP000076408
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9J7L6
A0A3S2NLQ9
A0A194PQA9
A0A194QSR4
A0A212FDA4
A0A2H1VM74
+ More
A0A2A4IWD7 A0A2A4IY13 A0A2A4IXZ9 A0A2A4IXA9 A0A026VX34 E2AXA5 A0A1B6MQ73 E2BI82 A0A1B6JTY9 A0A224XLZ6 A0A069DV06 A0A154PJT0 A0A0P4VZC0 K7J9U3 A0A0J7KN29 A0A0L7QNF6 B4HAC6 A0A067RJM7 E9IEC9 Q2LZB0 A0A195EZR2 A0A1B6CV28 A0A0M8ZSK0 A0A3B0JSH9 F4W8X3 A0A0M4EKG4 A0A158NP16 D6WVB0 A0A195BZG9 A0A2J7R740 A0A195DWU7 A0A2R7WCV1 V5G6G8 B4HUI4 A0A0J9RPD7 A0A2H8TG73 B3M3M7 B4MLR7 U5EW21 J9JKZ4 A0A1L8DZI0 A0A1Y1K7F6 A0A1B0DBI0 Q9VRP4 Q29QT3 A0A1J1IJS8 A0A2J7R728 B4PIW7 B4L992 B0WPT1 T1HTP5 B4LBS1 A0A151X8D2 A0A2A3EKI5 A0A0K8UNB4 A0A1L8DZI6 B4J1J1 A0A2S2QFP9 A0A1W4UX39 J3JW50 B3NC85 A0A1Q3FCS7 A0A0L0CKV1 A0A034WGW8 A0A1B0CL29 A0A182K709 A0A182WK79 A0A182RQ97 A0A1I8P5V1 A0A0K8V5G1 Q0IFZ2 A0A182PN98 N6SZ54 U4US73 W8BVL2 A0A182LJP9 E0V974 Q1HRE4 A0A182LVZ9 A0A182HM55 A0A182TDR7 A0A1I8MTH9 A0A182XCS9 A0A1B6GVX8 A0A182VB58 A0A293M6R1 A0A182GQK9 A0A084W2U6 A0A1B0AQC7 A0A1A9XIM1 Q7Q774 A0A2R5L8N1 A0A182J233 A0A182Y0U4
A0A2A4IWD7 A0A2A4IY13 A0A2A4IXZ9 A0A2A4IXA9 A0A026VX34 E2AXA5 A0A1B6MQ73 E2BI82 A0A1B6JTY9 A0A224XLZ6 A0A069DV06 A0A154PJT0 A0A0P4VZC0 K7J9U3 A0A0J7KN29 A0A0L7QNF6 B4HAC6 A0A067RJM7 E9IEC9 Q2LZB0 A0A195EZR2 A0A1B6CV28 A0A0M8ZSK0 A0A3B0JSH9 F4W8X3 A0A0M4EKG4 A0A158NP16 D6WVB0 A0A195BZG9 A0A2J7R740 A0A195DWU7 A0A2R7WCV1 V5G6G8 B4HUI4 A0A0J9RPD7 A0A2H8TG73 B3M3M7 B4MLR7 U5EW21 J9JKZ4 A0A1L8DZI0 A0A1Y1K7F6 A0A1B0DBI0 Q9VRP4 Q29QT3 A0A1J1IJS8 A0A2J7R728 B4PIW7 B4L992 B0WPT1 T1HTP5 B4LBS1 A0A151X8D2 A0A2A3EKI5 A0A0K8UNB4 A0A1L8DZI6 B4J1J1 A0A2S2QFP9 A0A1W4UX39 J3JW50 B3NC85 A0A1Q3FCS7 A0A0L0CKV1 A0A034WGW8 A0A1B0CL29 A0A182K709 A0A182WK79 A0A182RQ97 A0A1I8P5V1 A0A0K8V5G1 Q0IFZ2 A0A182PN98 N6SZ54 U4US73 W8BVL2 A0A182LJP9 E0V974 Q1HRE4 A0A182LVZ9 A0A182HM55 A0A182TDR7 A0A1I8MTH9 A0A182XCS9 A0A1B6GVX8 A0A182VB58 A0A293M6R1 A0A182GQK9 A0A084W2U6 A0A1B0AQC7 A0A1A9XIM1 Q7Q774 A0A2R5L8N1 A0A182J233 A0A182Y0U4
PDB
2F6R
E-value=7.39155e-39,
Score=404
Ontologies
PATHWAY
GO
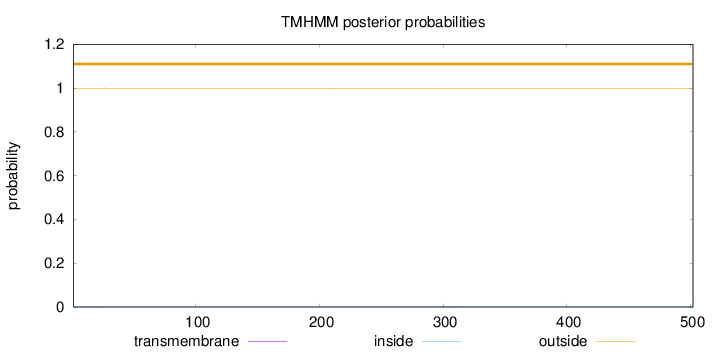
Topology
Length:
502
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00961999999999997
Exp number, first 60 AAs:
0.00517
Total prob of N-in:
0.00075
outside
1 - 502
Population Genetic Test Statistics
Pi
174.281061
Theta
204.179956
Tajima's D
-0.877566
CLR
0.529933
CSRT
0.15949202539873
Interpretation
Possibly Positive selection
