Gene
KWMTBOMO04646
Pre Gene Modal
BGIBMGA005507
Annotation
methuselah-like_protein_10_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.85
Sequence
CDS
ATGTCAATATACAACAATTCGGTTGCGTCCTCGCTGGTGGCTGCCGTCTGCTACCCTGACAGCGTTAGCAGCGACGACAACCACACGCTTTACATCGCGTATGCCGTTGGTCTTCTCCTGTCCGTGCCTTTTCTCCTCGCTACGTTCCTGGTATACGCCATGATCCCTGAGCTGAGGAACCTACACGGCATGTGTCTGATGGCGTATTGCGCGGGATTAATCGTCGGCTACCCCTTCCTTTCGTACCTGAAGCTGCACGTTGGCAGAATCGGAGCCAATATGACAGGATGCATTGTGTCTGCTTTTTTCGTGTACTACGCGTTTCAAACAACTTTCTTCTGGCTAAACGTAATGTGCTTCGATATATGGAGGACTTTCAGCGGTTATCGTGGTGGTAGCATGAACAAGCGCCGTGAAACGCGAAGCTGA
Protein
MSIYNNSVASSLVAAVCYPDSVSSDDNHTLYIAYAVGLLLSVPFLLATFLVYAMIPELRNLHGMCLMAYCAGLIVGYPFLSYLKLHVGRIGANMTGCIVSAFFVYYAFQTTFFWLNVMCFDIWRTFSGYRGGSMNKRRETRS
Summary
Similarity
Belongs to the G-protein coupled receptor Fz/Smo family.
Uniprot
J7EQT7
A0A2H1VHB6
A0A0L7LE48
A0A1E1WH08
A0A1E1WCB0
A0A194QSV3
+ More
A0A1E1WN64 A0A212FD97 H9J7L5 A0A194PWW7 A0A2A4JMU2 A0A2J7R7H5 A0A1L8DK28 A0A1L8DJU0 A0A1Q3FKC1 A0A1Q3FKG5 B0X3W8 A0A023ESZ7 A0A336MQY2 A0A336LUJ7 A0A336LZ61 A0A336MLF9 A0A336M665 A0A336MK75 A0A336LU04 A0A336LXU4 A0A182GN29 A0A182IR24 E2AKW8 A0A1B0CCQ5 A0A2S2NZ64 A0A3L8D8G7 E0VM50 K7JQP3 A0A154NYM6 A0A182TDZ9 A0A151X6P0 A0A1B6IUT9 A0A1B6IBM7 A0A2Y9D3K1 A0A182I170 A0A2Y9D3M1 A0A182V7J0 J7ETA9 Q7PNT9 A0A182LLL5 A0A0N0BJ15 A0A182FDT7 A0A1B6KQY4 A0A182YKX8 A0A195EHH8 A0A158NSZ5 Q16RE7 A0A182Q5Y6 A0A0C9S185 A0A182N958 A0A151I3Y4 A0A1B6JBW8 A0A1B6DQS8 A0A182PLA5 A0A182M5Z5 A0A182WLQ8 A0A195CCU5 A0A1B6F3W1 F4WY56 A0A182LY68 A0A1B6E0C8 A0A067QSF7 A0A2H8TDW7 A0A182SZW2 A0A1B6CU80 A0A1B6LT28 W5JCY9 A0A1B6G5F1 A0A195FEV0 A0A1B6F351 A0A182MPY5 A0A182KGA8 J9JKN2 A0A1W4W0K3 A0S6W9 A0A1W4VN78 A0A1W4W191 A0A1W4VNG3 A0A1W4VNF8 A0A1J1IGW7 A0A084WK39 A0A2P8ZJ24 A0A182I172 A0A182FDQ8 E2AK97 A7UU57 A0A182V1K9 A0A2Y9D3T7 A0A182LLL3 A0A336LXX0 A0A182THK7
A0A1E1WN64 A0A212FD97 H9J7L5 A0A194PWW7 A0A2A4JMU2 A0A2J7R7H5 A0A1L8DK28 A0A1L8DJU0 A0A1Q3FKC1 A0A1Q3FKG5 B0X3W8 A0A023ESZ7 A0A336MQY2 A0A336LUJ7 A0A336LZ61 A0A336MLF9 A0A336M665 A0A336MK75 A0A336LU04 A0A336LXU4 A0A182GN29 A0A182IR24 E2AKW8 A0A1B0CCQ5 A0A2S2NZ64 A0A3L8D8G7 E0VM50 K7JQP3 A0A154NYM6 A0A182TDZ9 A0A151X6P0 A0A1B6IUT9 A0A1B6IBM7 A0A2Y9D3K1 A0A182I170 A0A2Y9D3M1 A0A182V7J0 J7ETA9 Q7PNT9 A0A182LLL5 A0A0N0BJ15 A0A182FDT7 A0A1B6KQY4 A0A182YKX8 A0A195EHH8 A0A158NSZ5 Q16RE7 A0A182Q5Y6 A0A0C9S185 A0A182N958 A0A151I3Y4 A0A1B6JBW8 A0A1B6DQS8 A0A182PLA5 A0A182M5Z5 A0A182WLQ8 A0A195CCU5 A0A1B6F3W1 F4WY56 A0A182LY68 A0A1B6E0C8 A0A067QSF7 A0A2H8TDW7 A0A182SZW2 A0A1B6CU80 A0A1B6LT28 W5JCY9 A0A1B6G5F1 A0A195FEV0 A0A1B6F351 A0A182MPY5 A0A182KGA8 J9JKN2 A0A1W4W0K3 A0S6W9 A0A1W4VN78 A0A1W4W191 A0A1W4VNG3 A0A1W4VNF8 A0A1J1IGW7 A0A084WK39 A0A2P8ZJ24 A0A182I172 A0A182FDQ8 E2AK97 A7UU57 A0A182V1K9 A0A2Y9D3T7 A0A182LLL3 A0A336LXX0 A0A182THK7
Pubmed
EMBL
HQ188201
AEM43036.1
ODYU01002552
SOQ40206.1
JTDY01001460
KOB73813.1
+ More
GDQN01004782 JAT86272.1 GDQN01006455 JAT84599.1 KQ461154 KPJ08557.1 GDQN01002620 JAT88434.1 AGBW02009085 OWR51711.1 BABH01014015 KQ459595 KPI95630.1 NWSH01000966 PCG73367.1 NEVH01006732 PNF36788.1 GFDF01007357 JAV06727.1 GFDF01007356 JAV06728.1 GFDL01007037 JAV28008.1 GFDL01007020 JAV28025.1 DS232324 EDS40045.1 GAPW01001180 JAC12418.1 UFQT01001465 SSX30677.1 UFQT01000196 SSX21530.1 SSX21527.1 SSX30675.1 SSX21528.1 SSX30676.1 SSX21526.1 SSX21529.1 JXUM01014371 JXUM01014372 JXUM01014373 JXUM01014374 JXUM01014375 JXUM01014376 JXUM01014377 JXUM01014378 KQ560401 KXJ82611.1 GL440425 EFN65885.1 AJWK01006988 AJWK01006989 GGMR01009795 MBY22414.1 QOIP01000011 RLU16797.1 DS235289 EEB14456.1 KQ434783 KZC04727.1 KQ982476 KYQ56046.1 GECU01017005 JAS90701.1 GECU01023436 JAS84270.1 APCN01000032 HQ188200 AEM43035.1 AAAB01008960 EAA11607.3 KQ435724 KOX78511.1 GEBQ01026124 JAT13853.1 KQ978957 KYN27327.1 ADTU01025461 ADTU01025462 CH477715 EAT36973.2 AXCN02000556 GBYB01014511 JAG84278.1 KQ976472 KYM83995.1 GECU01011109 GECU01002381 JAS96597.1 JAT05326.1 GEDC01018773 GEDC01009262 GEDC01000043 JAS18525.1 JAS28036.1 JAS37255.1 AXCM01019671 KQ978023 KYM98041.1 GECZ01024998 JAS44771.1 GL888439 EGI60813.1 AXCM01013309 AXCM01013310 GEDC01018212 GEDC01005937 GEDC01001428 JAS19086.1 JAS31361.1 JAS35870.1 KK853008 KDR12560.1 GFXV01000486 MBW12291.1 GEDC01020320 JAS16978.1 GEBQ01013134 JAT26843.1 ADMH02001746 ETN61223.1 GECZ01012105 JAS57664.1 KQ981636 KYN38891.1 GECZ01025080 JAS44689.1 AXCM01007268 ABLF02038161 DQ295917 ABC24708.1 CVRI01000050 CRK99459.1 ATLV01024093 KE525349 KFB50583.1 PYGN01000041 PSN56483.1 APCN01000034 GL440209 EFN66138.1 EDO63835.1 SSX21523.1
GDQN01004782 JAT86272.1 GDQN01006455 JAT84599.1 KQ461154 KPJ08557.1 GDQN01002620 JAT88434.1 AGBW02009085 OWR51711.1 BABH01014015 KQ459595 KPI95630.1 NWSH01000966 PCG73367.1 NEVH01006732 PNF36788.1 GFDF01007357 JAV06727.1 GFDF01007356 JAV06728.1 GFDL01007037 JAV28008.1 GFDL01007020 JAV28025.1 DS232324 EDS40045.1 GAPW01001180 JAC12418.1 UFQT01001465 SSX30677.1 UFQT01000196 SSX21530.1 SSX21527.1 SSX30675.1 SSX21528.1 SSX30676.1 SSX21526.1 SSX21529.1 JXUM01014371 JXUM01014372 JXUM01014373 JXUM01014374 JXUM01014375 JXUM01014376 JXUM01014377 JXUM01014378 KQ560401 KXJ82611.1 GL440425 EFN65885.1 AJWK01006988 AJWK01006989 GGMR01009795 MBY22414.1 QOIP01000011 RLU16797.1 DS235289 EEB14456.1 KQ434783 KZC04727.1 KQ982476 KYQ56046.1 GECU01017005 JAS90701.1 GECU01023436 JAS84270.1 APCN01000032 HQ188200 AEM43035.1 AAAB01008960 EAA11607.3 KQ435724 KOX78511.1 GEBQ01026124 JAT13853.1 KQ978957 KYN27327.1 ADTU01025461 ADTU01025462 CH477715 EAT36973.2 AXCN02000556 GBYB01014511 JAG84278.1 KQ976472 KYM83995.1 GECU01011109 GECU01002381 JAS96597.1 JAT05326.1 GEDC01018773 GEDC01009262 GEDC01000043 JAS18525.1 JAS28036.1 JAS37255.1 AXCM01019671 KQ978023 KYM98041.1 GECZ01024998 JAS44771.1 GL888439 EGI60813.1 AXCM01013309 AXCM01013310 GEDC01018212 GEDC01005937 GEDC01001428 JAS19086.1 JAS31361.1 JAS35870.1 KK853008 KDR12560.1 GFXV01000486 MBW12291.1 GEDC01020320 JAS16978.1 GEBQ01013134 JAT26843.1 ADMH02001746 ETN61223.1 GECZ01012105 JAS57664.1 KQ981636 KYN38891.1 GECZ01025080 JAS44689.1 AXCM01007268 ABLF02038161 DQ295917 ABC24708.1 CVRI01000050 CRK99459.1 ATLV01024093 KE525349 KFB50583.1 PYGN01000041 PSN56483.1 APCN01000034 GL440209 EFN66138.1 EDO63835.1 SSX21523.1
Proteomes
UP000037510
UP000053240
UP000007151
UP000005204
UP000053268
UP000218220
+ More
UP000235965 UP000002320 UP000069940 UP000249989 UP000075880 UP000000311 UP000092461 UP000279307 UP000009046 UP000002358 UP000076502 UP000075902 UP000075809 UP000076407 UP000075840 UP000075903 UP000005203 UP000007062 UP000075882 UP000053105 UP000069272 UP000076408 UP000078492 UP000005205 UP000008820 UP000075886 UP000075884 UP000078540 UP000075885 UP000075883 UP000075920 UP000078542 UP000007755 UP000027135 UP000075901 UP000000673 UP000078541 UP000075881 UP000007819 UP000192221 UP000183832 UP000030765 UP000245037
UP000235965 UP000002320 UP000069940 UP000249989 UP000075880 UP000000311 UP000092461 UP000279307 UP000009046 UP000002358 UP000076502 UP000075902 UP000075809 UP000076407 UP000075840 UP000075903 UP000005203 UP000007062 UP000075882 UP000053105 UP000069272 UP000076408 UP000078492 UP000005205 UP000008820 UP000075886 UP000075884 UP000078540 UP000075885 UP000075883 UP000075920 UP000078542 UP000007755 UP000027135 UP000075901 UP000000673 UP000078541 UP000075881 UP000007819 UP000192221 UP000183832 UP000030765 UP000245037
Interpro
SUPFAM
SSF63877
SSF63877
Gene 3D
CDD
ProteinModelPortal
J7EQT7
A0A2H1VHB6
A0A0L7LE48
A0A1E1WH08
A0A1E1WCB0
A0A194QSV3
+ More
A0A1E1WN64 A0A212FD97 H9J7L5 A0A194PWW7 A0A2A4JMU2 A0A2J7R7H5 A0A1L8DK28 A0A1L8DJU0 A0A1Q3FKC1 A0A1Q3FKG5 B0X3W8 A0A023ESZ7 A0A336MQY2 A0A336LUJ7 A0A336LZ61 A0A336MLF9 A0A336M665 A0A336MK75 A0A336LU04 A0A336LXU4 A0A182GN29 A0A182IR24 E2AKW8 A0A1B0CCQ5 A0A2S2NZ64 A0A3L8D8G7 E0VM50 K7JQP3 A0A154NYM6 A0A182TDZ9 A0A151X6P0 A0A1B6IUT9 A0A1B6IBM7 A0A2Y9D3K1 A0A182I170 A0A2Y9D3M1 A0A182V7J0 J7ETA9 Q7PNT9 A0A182LLL5 A0A0N0BJ15 A0A182FDT7 A0A1B6KQY4 A0A182YKX8 A0A195EHH8 A0A158NSZ5 Q16RE7 A0A182Q5Y6 A0A0C9S185 A0A182N958 A0A151I3Y4 A0A1B6JBW8 A0A1B6DQS8 A0A182PLA5 A0A182M5Z5 A0A182WLQ8 A0A195CCU5 A0A1B6F3W1 F4WY56 A0A182LY68 A0A1B6E0C8 A0A067QSF7 A0A2H8TDW7 A0A182SZW2 A0A1B6CU80 A0A1B6LT28 W5JCY9 A0A1B6G5F1 A0A195FEV0 A0A1B6F351 A0A182MPY5 A0A182KGA8 J9JKN2 A0A1W4W0K3 A0S6W9 A0A1W4VN78 A0A1W4W191 A0A1W4VNG3 A0A1W4VNF8 A0A1J1IGW7 A0A084WK39 A0A2P8ZJ24 A0A182I172 A0A182FDQ8 E2AK97 A7UU57 A0A182V1K9 A0A2Y9D3T7 A0A182LLL3 A0A336LXX0 A0A182THK7
A0A1E1WN64 A0A212FD97 H9J7L5 A0A194PWW7 A0A2A4JMU2 A0A2J7R7H5 A0A1L8DK28 A0A1L8DJU0 A0A1Q3FKC1 A0A1Q3FKG5 B0X3W8 A0A023ESZ7 A0A336MQY2 A0A336LUJ7 A0A336LZ61 A0A336MLF9 A0A336M665 A0A336MK75 A0A336LU04 A0A336LXU4 A0A182GN29 A0A182IR24 E2AKW8 A0A1B0CCQ5 A0A2S2NZ64 A0A3L8D8G7 E0VM50 K7JQP3 A0A154NYM6 A0A182TDZ9 A0A151X6P0 A0A1B6IUT9 A0A1B6IBM7 A0A2Y9D3K1 A0A182I170 A0A2Y9D3M1 A0A182V7J0 J7ETA9 Q7PNT9 A0A182LLL5 A0A0N0BJ15 A0A182FDT7 A0A1B6KQY4 A0A182YKX8 A0A195EHH8 A0A158NSZ5 Q16RE7 A0A182Q5Y6 A0A0C9S185 A0A182N958 A0A151I3Y4 A0A1B6JBW8 A0A1B6DQS8 A0A182PLA5 A0A182M5Z5 A0A182WLQ8 A0A195CCU5 A0A1B6F3W1 F4WY56 A0A182LY68 A0A1B6E0C8 A0A067QSF7 A0A2H8TDW7 A0A182SZW2 A0A1B6CU80 A0A1B6LT28 W5JCY9 A0A1B6G5F1 A0A195FEV0 A0A1B6F351 A0A182MPY5 A0A182KGA8 J9JKN2 A0A1W4W0K3 A0S6W9 A0A1W4VN78 A0A1W4W191 A0A1W4VNG3 A0A1W4VNF8 A0A1J1IGW7 A0A084WK39 A0A2P8ZJ24 A0A182I172 A0A182FDQ8 E2AK97 A7UU57 A0A182V1K9 A0A2Y9D3T7 A0A182LLL3 A0A336LXX0 A0A182THK7
Ontologies
GO
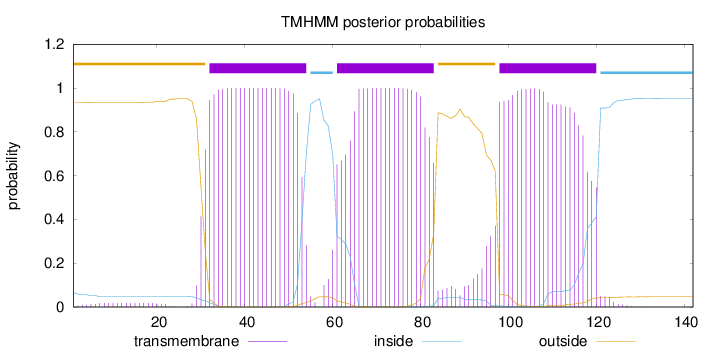
Topology
Length:
142
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
67.41153
Exp number, first 60 AAs:
23.76196
Total prob of N-in:
0.06543
POSSIBLE N-term signal
sequence
outside
1 - 31
TMhelix
32 - 54
inside
55 - 60
TMhelix
61 - 83
outside
84 - 97
TMhelix
98 - 120
inside
121 - 142
Population Genetic Test Statistics
Pi
180.772017
Theta
155.581475
Tajima's D
0.3153
CLR
0.459111
CSRT
0.462826858657067
Interpretation
Uncertain
