Gene
KWMTBOMO04640
Annotation
PREDICTED:_methylated-DNA--protein-cysteine_methyltransferase_isoform_X1_[Bombyx_mori]
Full name
Probable methylated-DNA--protein-cysteine methyltransferase
+ More
Methylated-DNA--protein-cysteine methyltransferase
Methylated-DNA--protein-cysteine methyltransferase
Alternative Name
6-O-methylguanine-DNA methyltransferase
O-6-methylguanine-DNA-alkyltransferase
O-6-methylguanine-DNA-alkyltransferase
Location in the cell
Mitochondrial Reliability : 2.409
Sequence
CDS
ATGAAATTACAAGAAATTCCCTTTGGCTCCACTTATACATATAGTGACATTACTAAGGCATTGGGACGACCAACAACCCATGCTAGAGCAGTAGGTTCAGCCTGTGGTGCCAATACACATCTTATAGTTACTCCATGTCATAGATTAATTAGTTTATCTTCTAAAGGTGGATTTAATAGTGGAATCAATCGTAAAGAATGGCTTATTGAACATGAGAAGAAATATGCAAACAATCTATGA
Protein
MKLQEIPFGSTYTYSDITKALGRPTTHARAVGSACGANTHLIVTPCHRLISLSSKGGFNSGINRKEWLIEHEKKYANNL
Summary
Description
Involved in the cellular defense against the biological effects of O6-methylguanine (O6-MeG) and O4-methylthymine (O4-MeT) in DNA. Repairs the methylated nucleobase in DNA by stoichiometrically transferring the methyl group to a cysteine residue in the enzyme. This is a suicide reaction: the enzyme is irreversibly inactivated.
Catalytic Activity
a 6-O-methyl-2'-deoxyguanosine in DNA + L-cysteinyl-[protein] = a 2'-deoxyguanosine in DNA + S-methyl-L-cysteinyl-[protein]
a 4-O-methyl-thymidine in DNA + L-cysteinyl-[protein] = a thymidine in DNA + S-methyl-L-cysteinyl-[protein]
a 4-O-methyl-thymidine in DNA + L-cysteinyl-[protein] = a thymidine in DNA + S-methyl-L-cysteinyl-[protein]
Cofactor
Zn(2+)
Miscellaneous
This enzyme catalyzes only one turnover and therefore is not strictly catalytic. According to one definition, an enzyme is a biocatalyst that acts repeatedly and over many reaction cycles.
Similarity
Belongs to the MGMT family.
Keywords
Complete proteome
DNA damage
DNA repair
Methyltransferase
Reference proteome
Transferase
Feature
chain Probable methylated-DNA--protein-cysteine methyltransferase
Uniprot
A0A194PQA4
A0A194QY97
A0A3S2NH25
A0A2H1VHG3
S4PBA3
A0A212FQ31
+ More
A0A2A4JPN0 A0A1Q3W732 B3ER72 V5L2G8 A0A159ZQS5 A0A0U2K3R1 J2YBN4 A0A140E125 A0A1E1EU91 A0A0G2Y1B6 Q5UNU9 A0A0R2H085 A0A2A3HVM5 A0A3S0ZCS3 A0A2V8TG66 A0A1I0URB1 A0A2A2IHY2 A0A2U3BW07 A0A3N5UXR7 A0A0G2Y3R2 A0A0C2LU67 H8LPL4 A0A2A4FKQ0 A0A2T5RIP0 A0A2P1EL10 G0GYQ3 A0A0F3RCJ1 A0A3B7DB19 A0A2A8VC95 A0A0P4UTG9 A0A2E2TI68 A0A1H1FJM1 A0A0F3QBW9 A0A0F3QIH7 A0A1S5V092 A0A1Q4RVY6 Q1RIP0 A0A1C3EW44 A0A249E1N8 A0A124R6V0 A0A1R1JCR7 A0A0M5JGE0 A0A0B2FAT4 A0A117XBQ5 A0A3G6F101 H2EF51 A0A1Z4QWW2 A0A2P1PA98 A0A167R6J9 A0A127W461 E4Q357 K2BQY8 A0A382HFY1 A0A1R1HZ59 A0A398DLM1 A0A346DEA3 A0A316GLT5 M5QSV9 A0A1G5S5T3 A0A0D0RSG7 A0A0D0G975 A0A1X6MAK1 A0A1W1XNI1 A0A348WH92 A0A257QQL4 A0A2A4HV95 A0A2A3T0Z1 A0A398D5I4 A0A2V8BSB2 A0A1M3H0B0 A0A0J0YUH7 A0A0A8X0G3 A0A348PSG1 A0A259DDP9 A0A2H5AWJ8 A0A2G8REY0 A0A3N5J5V3 Q12TZ2 A0A3Q9B8H5 A0A0C2FQ74 A0A0B1QC13 A0A1G0G7Y3 A0A1B9AHG7 A0A2E2U192 A0A2V8D7D0 A0A3N5JI11 A0A3D8PJW4 A0A3M1YBR3 B9MNR0 A0A268J0X3 A0A1G0IRE6 A0A2G1YJQ1 A0A0K6GKE3 A0A081NXJ7
A0A2A4JPN0 A0A1Q3W732 B3ER72 V5L2G8 A0A159ZQS5 A0A0U2K3R1 J2YBN4 A0A140E125 A0A1E1EU91 A0A0G2Y1B6 Q5UNU9 A0A0R2H085 A0A2A3HVM5 A0A3S0ZCS3 A0A2V8TG66 A0A1I0URB1 A0A2A2IHY2 A0A2U3BW07 A0A3N5UXR7 A0A0G2Y3R2 A0A0C2LU67 H8LPL4 A0A2A4FKQ0 A0A2T5RIP0 A0A2P1EL10 G0GYQ3 A0A0F3RCJ1 A0A3B7DB19 A0A2A8VC95 A0A0P4UTG9 A0A2E2TI68 A0A1H1FJM1 A0A0F3QBW9 A0A0F3QIH7 A0A1S5V092 A0A1Q4RVY6 Q1RIP0 A0A1C3EW44 A0A249E1N8 A0A124R6V0 A0A1R1JCR7 A0A0M5JGE0 A0A0B2FAT4 A0A117XBQ5 A0A3G6F101 H2EF51 A0A1Z4QWW2 A0A2P1PA98 A0A167R6J9 A0A127W461 E4Q357 K2BQY8 A0A382HFY1 A0A1R1HZ59 A0A398DLM1 A0A346DEA3 A0A316GLT5 M5QSV9 A0A1G5S5T3 A0A0D0RSG7 A0A0D0G975 A0A1X6MAK1 A0A1W1XNI1 A0A348WH92 A0A257QQL4 A0A2A4HV95 A0A2A3T0Z1 A0A398D5I4 A0A2V8BSB2 A0A1M3H0B0 A0A0J0YUH7 A0A0A8X0G3 A0A348PSG1 A0A259DDP9 A0A2H5AWJ8 A0A2G8REY0 A0A3N5J5V3 Q12TZ2 A0A3Q9B8H5 A0A0C2FQ74 A0A0B1QC13 A0A1G0G7Y3 A0A1B9AHG7 A0A2E2U192 A0A2V8D7D0 A0A3N5JI11 A0A3D8PJW4 A0A3M1YBR3 B9MNR0 A0A268J0X3 A0A1G0IRE6 A0A2G1YJQ1 A0A0K6GKE3 A0A081NXJ7
EC Number
2.1.1.63
Pubmed
EMBL
KQ459595
KPI95626.1
KQ461154
KPJ08551.1
RSAL01000041
RVE50878.1
+ More
ODYU01002578 SOQ40267.1 GAIX01006012 JAA86548.1 AGBW02000285 OWR55799.1 NWSH01000966 PCG73372.1 MKUM01000080 OJW68423.1 CP001102 ACE05724.1 KF493731 AHA45142.1 KU761889 AMZ03137.1 KT599914 ALR84315.1 AFYC01000005 EJN41102.1 KF959826 AMK62000.1 AP017644 AP017645 BAV61823.1 BAV62809.1 HQ336222 JN036606 KM982401 KM982403 ADO18794.1 AEJ34935.1 AKI79480.1 AKI81371.1 AY653733 JQBM01000002 OBHM01000005 KRN46287.1 SOB43214.1 NSGV01000002 PBC71782.1 RZJR01000001 RUS48981.1 QHXT01000021 PYT10260.1 FOJP01000022 SFA66599.1 NPOA01000001 PAV31611.1 NSKH01000024 PWI40960.1 RPPI01000117 RPJ26849.1 KM982402 AKI80433.1 JXCB01000004 KIJ78381.1 CP003375 AFD19879.1 MTZU01000020 PCE33238.1 QAXS01000018 PTV98217.1 MG807320 AVL94582.1 CP002912 AEK74861.1 LAOQ01000007 KJW04008.1 CP032049 AXU06696.1 NTUP01000013 PFA67722.1 BBWW01000001 GAP94866.1 PAMH01000008 MAZ38797.1 FNKD01000004 SDR01242.1 LAOI01000001 KJV89646.1 LAOJ01000001 KJV91961.1 KY110734 AQN68063.1 MRCD01000017 OKH52954.1 CP000087 ABE04774.1 LYMV01000121 ODA37617.1 CP016305 ASX27435.1 LOXM01000279 KVG53106.1 MTJZ01000012 OMG73157.1 CP012602 ALC92157.1 JUEI01000015 KHL96208.1 LOTN01000055 KUZ84541.1 CP033854 AYZ62163.1 JN885999 AEX63116.1 AP018280 BAZ42410.1 CP027845 AVP88188.1 KU877344 ANB50366.1 CP014616 AMQ08406.1 CP002216 ADQ03893.1 AMFJ01019640 EKD71303.1 UINC01060997 SVB86099.1 MTHC01000027 OMG51752.1 QXIW01000027 RIE13017.1 CP025535 AXM88431.1 QGGW01000003 PWK60879.1 ANMT01000020 JRZG01000007 EMI10325.1 KGP61385.1 FMWL01000022 SCZ81772.1 JXTH01000020 KIQ94597.1 JXTG01000003 KIP21885.1 NDEZ01000043 OSX53398.1 FWXF01000013 SMC25405.1 DMVW01000179 HAR53904.1 NCAP01000104 OYV50086.1 NWVD01000009 PCG07943.1 NVQX01000015 PBO81709.1 QXIT01000093 RIE07698.1 QHWH01000124 PYQ99003.1 MKUL01000037 OJW51112.1 JTDO01000002 KLT73749.1 BASE01000034 GAM13455.1 DMTL01000216 HAQ71722.1 NCHW01000478 OZA12089.1 CP025394 AUG76529.1 AWWI01000066 PIL20144.1 RPQS01000213 RPH92200.1 CP000300 ABE53084.1 CP032760 AZO96742.1 KN749036 KIH50670.1 JRFJ01000001 KHJ56370.1 MGXP01000020 OGT33719.1 MAYU01000023 OCA83288.1 PANA01000005 MAZ45033.1 QHWJ01000179 QHWI01000318 PYR21023.1 PYR46911.1 RPQS01000049 RPH95193.1 PIOD01000025 RDW15519.1 RFHL01000031 RMG75660.1 CP001393 ACM59589.1 NPDE01000068 PAE28035.1 MGYI01000155 OGT64723.1 NVRA01000043 PHQ63752.1 CYGZ01000003 CUA79204.1 JNVM01000024 KEQ23170.1
ODYU01002578 SOQ40267.1 GAIX01006012 JAA86548.1 AGBW02000285 OWR55799.1 NWSH01000966 PCG73372.1 MKUM01000080 OJW68423.1 CP001102 ACE05724.1 KF493731 AHA45142.1 KU761889 AMZ03137.1 KT599914 ALR84315.1 AFYC01000005 EJN41102.1 KF959826 AMK62000.1 AP017644 AP017645 BAV61823.1 BAV62809.1 HQ336222 JN036606 KM982401 KM982403 ADO18794.1 AEJ34935.1 AKI79480.1 AKI81371.1 AY653733 JQBM01000002 OBHM01000005 KRN46287.1 SOB43214.1 NSGV01000002 PBC71782.1 RZJR01000001 RUS48981.1 QHXT01000021 PYT10260.1 FOJP01000022 SFA66599.1 NPOA01000001 PAV31611.1 NSKH01000024 PWI40960.1 RPPI01000117 RPJ26849.1 KM982402 AKI80433.1 JXCB01000004 KIJ78381.1 CP003375 AFD19879.1 MTZU01000020 PCE33238.1 QAXS01000018 PTV98217.1 MG807320 AVL94582.1 CP002912 AEK74861.1 LAOQ01000007 KJW04008.1 CP032049 AXU06696.1 NTUP01000013 PFA67722.1 BBWW01000001 GAP94866.1 PAMH01000008 MAZ38797.1 FNKD01000004 SDR01242.1 LAOI01000001 KJV89646.1 LAOJ01000001 KJV91961.1 KY110734 AQN68063.1 MRCD01000017 OKH52954.1 CP000087 ABE04774.1 LYMV01000121 ODA37617.1 CP016305 ASX27435.1 LOXM01000279 KVG53106.1 MTJZ01000012 OMG73157.1 CP012602 ALC92157.1 JUEI01000015 KHL96208.1 LOTN01000055 KUZ84541.1 CP033854 AYZ62163.1 JN885999 AEX63116.1 AP018280 BAZ42410.1 CP027845 AVP88188.1 KU877344 ANB50366.1 CP014616 AMQ08406.1 CP002216 ADQ03893.1 AMFJ01019640 EKD71303.1 UINC01060997 SVB86099.1 MTHC01000027 OMG51752.1 QXIW01000027 RIE13017.1 CP025535 AXM88431.1 QGGW01000003 PWK60879.1 ANMT01000020 JRZG01000007 EMI10325.1 KGP61385.1 FMWL01000022 SCZ81772.1 JXTH01000020 KIQ94597.1 JXTG01000003 KIP21885.1 NDEZ01000043 OSX53398.1 FWXF01000013 SMC25405.1 DMVW01000179 HAR53904.1 NCAP01000104 OYV50086.1 NWVD01000009 PCG07943.1 NVQX01000015 PBO81709.1 QXIT01000093 RIE07698.1 QHWH01000124 PYQ99003.1 MKUL01000037 OJW51112.1 JTDO01000002 KLT73749.1 BASE01000034 GAM13455.1 DMTL01000216 HAQ71722.1 NCHW01000478 OZA12089.1 CP025394 AUG76529.1 AWWI01000066 PIL20144.1 RPQS01000213 RPH92200.1 CP000300 ABE53084.1 CP032760 AZO96742.1 KN749036 KIH50670.1 JRFJ01000001 KHJ56370.1 MGXP01000020 OGT33719.1 MAYU01000023 OCA83288.1 PANA01000005 MAZ45033.1 QHWJ01000179 QHWI01000318 PYR21023.1 PYR46911.1 RPQS01000049 RPH95193.1 PIOD01000025 RDW15519.1 RFHL01000031 RMG75660.1 CP001393 ACM59589.1 NPDE01000068 PAE28035.1 MGYI01000155 OGT64723.1 NVRA01000043 PHQ63752.1 CYGZ01000003 CUA79204.1 JNVM01000024 KEQ23170.1
Proteomes
UP000053268
UP000053240
UP000283053
UP000007151
UP000218220
UP000001227
+ More
UP000240740 UP000241559 UP000241834 UP000007297 UP000240935 UP000240366 UP000241484 UP000201519 UP000240552 UP000241474 UP000274448 UP000001134 UP000051992 UP000218662 UP000267180 UP000182453 UP000218887 UP000245673 UP000280678 UP000240461 UP000031981 UP000007592 UP000217994 UP000244089 UP000289600 UP000000865 UP000033736 UP000258667 UP000220634 UP000052243 UP000229611 UP000199444 UP000033661 UP000033689 UP000240801 UP000185789 UP000001951 UP000094749 UP000215668 UP000064029 UP000187194 UP000060713 UP000031063 UP000065521 UP000280242 UP000290974 UP000241365 UP000071057 UP000006889 UP000187142 UP000256325 UP000245708 UP000011915 UP000030117 UP000199208 UP000032102 UP000032047 UP000194126 UP000192783 UP000216804 UP000218784 UP000247608 UP000036027 UP000031014 UP000264021 UP000239175 UP000231259 UP000276319 UP000001979 UP000268874 UP000030826 UP000092646 UP000226955 UP000248064 UP000248353 UP000256520 UP000007723 UP000215893 UP000182738 UP000028123
UP000240740 UP000241559 UP000241834 UP000007297 UP000240935 UP000240366 UP000241484 UP000201519 UP000240552 UP000241474 UP000274448 UP000001134 UP000051992 UP000218662 UP000267180 UP000182453 UP000218887 UP000245673 UP000280678 UP000240461 UP000031981 UP000007592 UP000217994 UP000244089 UP000289600 UP000000865 UP000033736 UP000258667 UP000220634 UP000052243 UP000229611 UP000199444 UP000033661 UP000033689 UP000240801 UP000185789 UP000001951 UP000094749 UP000215668 UP000064029 UP000187194 UP000060713 UP000031063 UP000065521 UP000280242 UP000290974 UP000241365 UP000071057 UP000006889 UP000187142 UP000256325 UP000245708 UP000011915 UP000030117 UP000199208 UP000032102 UP000032047 UP000194126 UP000192783 UP000216804 UP000218784 UP000247608 UP000036027 UP000031014 UP000264021 UP000239175 UP000231259 UP000276319 UP000001979 UP000268874 UP000030826 UP000092646 UP000226955 UP000248064 UP000248353 UP000256520 UP000007723 UP000215893 UP000182738 UP000028123
Pfam
Interpro
IPR008332
MethylG_MeTrfase_N
+ More
IPR036217 MethylDNA_cys_MeTrfase_DNAb
IPR036631 MGMT_N_sf
IPR014048 MethylDNA_cys_MeTrfase_DNA-bd
IPR036388 WH-like_DNA-bd_sf
IPR001497 MethylDNA_cys_MeTrfase_AS
IPR041694 ADH_N_2
IPR036291 NAD(P)-bd_dom_sf
IPR013149 ADH_C
IPR011032 GroES-like_sf
IPR014190 PTGR1
IPR020843 PKS_ER
IPR016221 Bifunct_regulatory_prot_Ada
IPR009057 Homeobox-like_sf
IPR004026 Ada_DNA_repair_Zn-bd
IPR035451 Ada-like_dom_sf
IPR018060 HTH_AraC
IPR023546 MGMT
IPR011257 DNA_glycosylase
IPR003265 HhH-GPD_domain
IPR036217 MethylDNA_cys_MeTrfase_DNAb
IPR036631 MGMT_N_sf
IPR014048 MethylDNA_cys_MeTrfase_DNA-bd
IPR036388 WH-like_DNA-bd_sf
IPR001497 MethylDNA_cys_MeTrfase_AS
IPR041694 ADH_N_2
IPR036291 NAD(P)-bd_dom_sf
IPR013149 ADH_C
IPR011032 GroES-like_sf
IPR014190 PTGR1
IPR020843 PKS_ER
IPR016221 Bifunct_regulatory_prot_Ada
IPR009057 Homeobox-like_sf
IPR004026 Ada_DNA_repair_Zn-bd
IPR035451 Ada-like_dom_sf
IPR018060 HTH_AraC
IPR023546 MGMT
IPR011257 DNA_glycosylase
IPR003265 HhH-GPD_domain
SUPFAM
Gene 3D
ProteinModelPortal
A0A194PQA4
A0A194QY97
A0A3S2NH25
A0A2H1VHG3
S4PBA3
A0A212FQ31
+ More
A0A2A4JPN0 A0A1Q3W732 B3ER72 V5L2G8 A0A159ZQS5 A0A0U2K3R1 J2YBN4 A0A140E125 A0A1E1EU91 A0A0G2Y1B6 Q5UNU9 A0A0R2H085 A0A2A3HVM5 A0A3S0ZCS3 A0A2V8TG66 A0A1I0URB1 A0A2A2IHY2 A0A2U3BW07 A0A3N5UXR7 A0A0G2Y3R2 A0A0C2LU67 H8LPL4 A0A2A4FKQ0 A0A2T5RIP0 A0A2P1EL10 G0GYQ3 A0A0F3RCJ1 A0A3B7DB19 A0A2A8VC95 A0A0P4UTG9 A0A2E2TI68 A0A1H1FJM1 A0A0F3QBW9 A0A0F3QIH7 A0A1S5V092 A0A1Q4RVY6 Q1RIP0 A0A1C3EW44 A0A249E1N8 A0A124R6V0 A0A1R1JCR7 A0A0M5JGE0 A0A0B2FAT4 A0A117XBQ5 A0A3G6F101 H2EF51 A0A1Z4QWW2 A0A2P1PA98 A0A167R6J9 A0A127W461 E4Q357 K2BQY8 A0A382HFY1 A0A1R1HZ59 A0A398DLM1 A0A346DEA3 A0A316GLT5 M5QSV9 A0A1G5S5T3 A0A0D0RSG7 A0A0D0G975 A0A1X6MAK1 A0A1W1XNI1 A0A348WH92 A0A257QQL4 A0A2A4HV95 A0A2A3T0Z1 A0A398D5I4 A0A2V8BSB2 A0A1M3H0B0 A0A0J0YUH7 A0A0A8X0G3 A0A348PSG1 A0A259DDP9 A0A2H5AWJ8 A0A2G8REY0 A0A3N5J5V3 Q12TZ2 A0A3Q9B8H5 A0A0C2FQ74 A0A0B1QC13 A0A1G0G7Y3 A0A1B9AHG7 A0A2E2U192 A0A2V8D7D0 A0A3N5JI11 A0A3D8PJW4 A0A3M1YBR3 B9MNR0 A0A268J0X3 A0A1G0IRE6 A0A2G1YJQ1 A0A0K6GKE3 A0A081NXJ7
A0A2A4JPN0 A0A1Q3W732 B3ER72 V5L2G8 A0A159ZQS5 A0A0U2K3R1 J2YBN4 A0A140E125 A0A1E1EU91 A0A0G2Y1B6 Q5UNU9 A0A0R2H085 A0A2A3HVM5 A0A3S0ZCS3 A0A2V8TG66 A0A1I0URB1 A0A2A2IHY2 A0A2U3BW07 A0A3N5UXR7 A0A0G2Y3R2 A0A0C2LU67 H8LPL4 A0A2A4FKQ0 A0A2T5RIP0 A0A2P1EL10 G0GYQ3 A0A0F3RCJ1 A0A3B7DB19 A0A2A8VC95 A0A0P4UTG9 A0A2E2TI68 A0A1H1FJM1 A0A0F3QBW9 A0A0F3QIH7 A0A1S5V092 A0A1Q4RVY6 Q1RIP0 A0A1C3EW44 A0A249E1N8 A0A124R6V0 A0A1R1JCR7 A0A0M5JGE0 A0A0B2FAT4 A0A117XBQ5 A0A3G6F101 H2EF51 A0A1Z4QWW2 A0A2P1PA98 A0A167R6J9 A0A127W461 E4Q357 K2BQY8 A0A382HFY1 A0A1R1HZ59 A0A398DLM1 A0A346DEA3 A0A316GLT5 M5QSV9 A0A1G5S5T3 A0A0D0RSG7 A0A0D0G975 A0A1X6MAK1 A0A1W1XNI1 A0A348WH92 A0A257QQL4 A0A2A4HV95 A0A2A3T0Z1 A0A398D5I4 A0A2V8BSB2 A0A1M3H0B0 A0A0J0YUH7 A0A0A8X0G3 A0A348PSG1 A0A259DDP9 A0A2H5AWJ8 A0A2G8REY0 A0A3N5J5V3 Q12TZ2 A0A3Q9B8H5 A0A0C2FQ74 A0A0B1QC13 A0A1G0G7Y3 A0A1B9AHG7 A0A2E2U192 A0A2V8D7D0 A0A3N5JI11 A0A3D8PJW4 A0A3M1YBR3 B9MNR0 A0A268J0X3 A0A1G0IRE6 A0A2G1YJQ1 A0A0K6GKE3 A0A081NXJ7
PDB
4WXC
E-value=4.47398e-11,
Score=156
Ontologies
GO
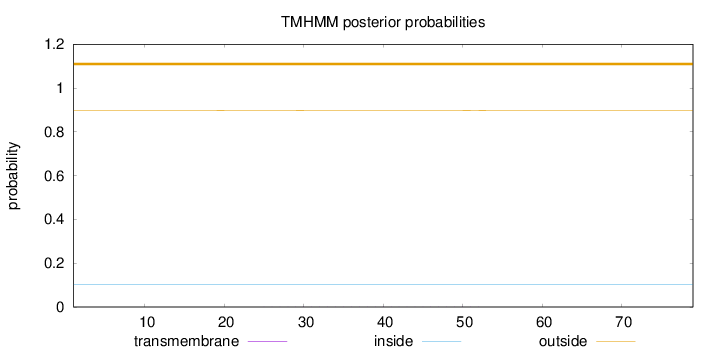
Topology
Subcellular location
Cytoplasm
Length:
79
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0163
Exp number, first 60 AAs:
0.01628
Total prob of N-in:
0.10176
outside
1 - 79
Population Genetic Test Statistics
Pi
76.548663
Theta
83.606199
Tajima's D
-0.264263
CLR
270.01167
CSRT
0.294335283235838
Interpretation
Uncertain
