Gene
KWMTBOMO04633
Pre Gene Modal
BGIBMGA005504
Annotation
PREDICTED:_transmembrane_protein_120A_isoform_X1_[Papilio_xuthus]
Full name
Transmembrane protein 120 homolog
Location in the cell
PlasmaMembrane Reliability : 3.455
Sequence
CDS
ATGGAAGTTGAAGAATGTGTGAAGGAATGGGAGGATTTAGTAGCAGATTACAAGCGCTTAGAGACTATCAATAAAGAGTACACAACAAAATTAGAAGAAGTTGGCGAACTGCAAGCAGCATGTTTGAAGGAACTGGAACACCAGAAATATCGCATGTCAATTATTAATGCTGCATTGAAAAGGTTGGAAAAGAAAGGCGGTGTGAAAGATGAACGCGTCACAAACCTCGAGAAGGAGATAATGAAGAGGAAAGCGATGCTTCACGAGATAAAAGCCACGCTCCCCAAACAGAACAGCTTATATCTGCGCATCATATTAGGAAACGTTAATGTATCTATATTAAACAAGAATGACAAATTTAAATACAAAGACGAGTATGAAAAATTCAAATTGATCCTCAGTGCGATCGCATTTATTTTAGCTGTAGGAAATCTGTATATTGACTTCAGGCCGCTGCAATTGATTTTAATTTTTCTTCTCGTCTGGTACTACTGCACGCTTACGATCCGAGAGAGTATACTTAAAGTGAACGGCTCAAGGATAAAAGGATGGTGGCGCTTACATCACTTCATATCGACGGTGGTTGCGGGAATATTGCTTATATGGCCCCAGAACCAGCCTTGGGAGGAATTTAGGCACACCTTCATGTGGTTTATTGCTTATATAAGTGTTGTACAATACATGCAGTTCAGGTACCAGAGTGGAGTACTATACAGGCTCAAAGCCTTGGGCGCCAGGCATAATATGGATATCACGATCGAAGGATTCCATTCGTGGATGTGGCGTGGTCTATCATATCTATTGCCTTTCTTGTTTGGGGGATATGTTTTTCAACTGTACATCGCTTATACATTGTACCATTTGAGTTACCATCCCGAGGCGACTTGGCAGGTGCCATATTTAGCTGCCTTGTTCCTTACTTTGCACTGTTGCAACATGTATACCATATTAAGGACATTACGCAGGAAGGTTAAAGGAGGCCTGAAACTGAGATACAAGTTGCGCGCCATTGCGTATCGGCTCTCCAATGAGATCGTCGTACTGGAACAAAAATGGCGGCAAAACAAAGTCGAACAGAAAGCGGAATAA
Protein
MEVEECVKEWEDLVADYKRLETINKEYTTKLEEVGELQAACLKELEHQKYRMSIINAALKRLEKKGGVKDERVTNLEKEIMKRKAMLHEIKATLPKQNSLYLRIILGNVNVSILNKNDKFKYKDEYEKFKLILSAIAFILAVGNLYIDFRPLQLILIFLLVWYYCTLTIRESILKVNGSRIKGWWRLHHFISTVVAGILLIWPQNQPWEEFRHTFMWFIAYISVVQYMQFRYQSGVLYRLKALGARHNMDITIEGFHSWMWRGLSYLLPFLFGGYVFQLYIAYTLYHLSYHPEATWQVPYLAALFLTLHCCNMYTILRTLRRKVKGGLKLRYKLRAIAYRLSNEIVVLEQKWRQNKVEQKAE
Summary
Similarity
Belongs to the TMEM120 family.
Keywords
Coiled coil
Complete proteome
Glycoprotein
Membrane
Phosphoprotein
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Transmembrane protein 120 homolog
Uniprot
H9J7L2
A0A2H1VHE9
A0A194PQN4
A0A194QSQ3
S4Q095
A0A1E1WNR6
+ More
A0A2A4IYH0 A0A2A4IXW2 A0A2J7Q8J3 A0A067RTI7 A0A2P8YR06 A0A195AVG2 A0A158NF48 A0A195E8C6 A0A0L7R8M0 A0A0M8ZSJ5 E9ID26 A0A026W2R5 A0A088APV8 E2B080 A0A2A3EI99 D6WH93 A0A1Y1N0H1 A0A0J7KY31 A0A1Q3FYR5 A0A0T6BE85 A0A1W4X4K8 A0A023EQX9 T1DEC3 A0A0K8TQ19 A0A195C425 A0A1B6L2L4 F4WEI9 A0A1B6HS06 E0VMX0 A0A182N8F2 A0A1D2NI48 T1E212 J3JUI3 A0A182Q6N7 A0A1L8DUU0 U4UKY9 N6SZG0 Q16NV2 A0A182U4M5 A0A182XBX6 A0A182UVS2 A0A182IAK7 K7IRZ8 Q7QF35 A0A1J1I9Q1 A0A2H8TRS2 A0A336MKB9 A0A2M4CI84 A0A2M4CIU0 A0A2M3Z7B0 B4JJE2 A0A084VNX3 A0A1B0B3X0 A0A182W5L5 A0A1B6DIN6 A0A2S2NFL9 A0A1A9XVI2 A0A2M3Z7E4 A0A1B6E3Y5 T1HQX1 A0A0P4VJF3 J9JQL9 D6X2J7 A0A069DSM7 A0A1A9W971 A0A226E637 A0A1A9V0K9 A0A182M9S0 D3TNX8 A0A1B0A8G5 A0A0V0G760 A0A182FIR7 A0A2M4AD83 A0A023FAD9 A0A182RHK2 T1DUG4 B4Q0V7 A0A1W4VS75 T1PC33 A0A2M4BRU8 A0A0M4ELC3 B3NTX3 B5DM77 A0A1W4VFI4 A0A1I8MKA9 A4V3W8 Q9U1M2 A0A162DGT0 B4R3U6 A0A0A1WFG4 A0A0P4YN02
A0A2A4IYH0 A0A2A4IXW2 A0A2J7Q8J3 A0A067RTI7 A0A2P8YR06 A0A195AVG2 A0A158NF48 A0A195E8C6 A0A0L7R8M0 A0A0M8ZSJ5 E9ID26 A0A026W2R5 A0A088APV8 E2B080 A0A2A3EI99 D6WH93 A0A1Y1N0H1 A0A0J7KY31 A0A1Q3FYR5 A0A0T6BE85 A0A1W4X4K8 A0A023EQX9 T1DEC3 A0A0K8TQ19 A0A195C425 A0A1B6L2L4 F4WEI9 A0A1B6HS06 E0VMX0 A0A182N8F2 A0A1D2NI48 T1E212 J3JUI3 A0A182Q6N7 A0A1L8DUU0 U4UKY9 N6SZG0 Q16NV2 A0A182U4M5 A0A182XBX6 A0A182UVS2 A0A182IAK7 K7IRZ8 Q7QF35 A0A1J1I9Q1 A0A2H8TRS2 A0A336MKB9 A0A2M4CI84 A0A2M4CIU0 A0A2M3Z7B0 B4JJE2 A0A084VNX3 A0A1B0B3X0 A0A182W5L5 A0A1B6DIN6 A0A2S2NFL9 A0A1A9XVI2 A0A2M3Z7E4 A0A1B6E3Y5 T1HQX1 A0A0P4VJF3 J9JQL9 D6X2J7 A0A069DSM7 A0A1A9W971 A0A226E637 A0A1A9V0K9 A0A182M9S0 D3TNX8 A0A1B0A8G5 A0A0V0G760 A0A182FIR7 A0A2M4AD83 A0A023FAD9 A0A182RHK2 T1DUG4 B4Q0V7 A0A1W4VS75 T1PC33 A0A2M4BRU8 A0A0M4ELC3 B3NTX3 B5DM77 A0A1W4VFI4 A0A1I8MKA9 A4V3W8 Q9U1M2 A0A162DGT0 B4R3U6 A0A0A1WFG4 A0A0P4YN02
Pubmed
19121390
26354079
23622113
24845553
29403074
21347285
+ More
21282665 24508170 30249741 20798317 18362917 19820115 28004739 24945155 24330624 26369729 21719571 20566863 27289101 22516182 23537049 17510324 20075255 12364791 17994087 24438588 27129103 26334808 20353571 25474469 17550304 15632085 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 10731137 12537569 18327897 25830018
21282665 24508170 30249741 20798317 18362917 19820115 28004739 24945155 24330624 26369729 21719571 20566863 27289101 22516182 23537049 17510324 20075255 12364791 17994087 24438588 27129103 26334808 20353571 25474469 17550304 15632085 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 10731137 12537569 18327897 25830018
EMBL
BABH01013994
ODYU01002578
SOQ40265.1
KQ459595
KPI95622.1
KQ461154
+ More
KPJ08548.1 GAIX01000178 JAA92382.1 GDQN01002410 JAT88644.1 NWSH01005291 PCG64314.1 PCG64316.1 NEVH01016952 PNF24904.1 KK852473 KDR23134.1 PYGN01000420 PSN46690.1 KQ976736 KYM76015.1 ADTU01013744 KQ979479 KYN21366.1 KQ414632 KOC67116.1 KQ435922 KOX68649.1 GL762454 EFZ21509.1 KK107522 QOIP01000001 EZA49324.1 RLU27293.1 GL444420 EFN60909.1 KZ288232 PBC31523.1 KQ971330 EFA00988.1 GEZM01016165 JAV91354.1 LBMM01002172 KMQ95184.1 GFDL01002326 JAV32719.1 LJIG01001286 KRT85654.1 GAPW01002070 JAC11528.1 GALA01001092 JAA93760.1 GDAI01001116 JAI16487.1 KQ978287 KYM95609.1 GEBQ01022096 JAT17881.1 GL888103 EGI67352.1 GECU01030233 JAS77473.1 DS235327 EEB14726.1 LJIJ01000033 ODN04921.1 GALA01001615 JAA93237.1 BT126897 AEE61859.1 AXCN02002153 GFDF01003886 JAV10198.1 KB632256 ERL90675.1 APGK01050872 KB741178 ENN73159.1 CH477807 EAT36034.1 APCN01001262 AAAB01008846 EAA06504.5 CVRI01000044 CRK96466.1 GFXV01004487 MBW16292.1 UFQS01001175 UFQS01001201 UFQT01001175 UFQT01001201 SSX09411.1 SSX09695.1 SSX29313.1 GGFL01000882 MBW65060.1 GGFL01000883 MBW65061.1 GGFM01003668 MBW24419.1 CH916370 EDV99694.1 ATLV01014939 ATLV01014940 KE524996 KFB39667.1 JXJN01008099 GEDC01023789 GEDC01022599 GEDC01011803 GEDC01011376 GEDC01008019 JAS13509.1 JAS14699.1 JAS25495.1 JAS25922.1 JAS29279.1 GGMR01003384 MBY16003.1 GGFM01003674 MBW24425.1 GEDC01004656 JAS32642.1 ACPB03027316 GDKW01002279 JAI54316.1 ABLF02022575 KQ971372 EFA09453.1 GBGD01001969 JAC86920.1 LNIX01000006 OXA52810.1 AXCM01004618 CCAG010008024 EZ423130 ADD19406.1 GECL01002232 JAP03892.1 GGFK01005420 MBW38741.1 GBBI01000271 JAC18441.1 GAMD01000049 JAB01542.1 CM000162 EDX01324.1 KA646249 AFP60878.1 GGFJ01006367 MBW55508.1 CP012528 ALC48196.1 CH954180 EDV45681.1 CH379064 EDY72594.1 AE014298 AAS65258.1 AFH07223.1 AHN59305.1 AL133506 AY118486 LRGB01001579 KZS11574.1 CM000366 EDX16958.1 GBXI01016876 JAC97415.1 GDIP01224987 JAI98414.1
KPJ08548.1 GAIX01000178 JAA92382.1 GDQN01002410 JAT88644.1 NWSH01005291 PCG64314.1 PCG64316.1 NEVH01016952 PNF24904.1 KK852473 KDR23134.1 PYGN01000420 PSN46690.1 KQ976736 KYM76015.1 ADTU01013744 KQ979479 KYN21366.1 KQ414632 KOC67116.1 KQ435922 KOX68649.1 GL762454 EFZ21509.1 KK107522 QOIP01000001 EZA49324.1 RLU27293.1 GL444420 EFN60909.1 KZ288232 PBC31523.1 KQ971330 EFA00988.1 GEZM01016165 JAV91354.1 LBMM01002172 KMQ95184.1 GFDL01002326 JAV32719.1 LJIG01001286 KRT85654.1 GAPW01002070 JAC11528.1 GALA01001092 JAA93760.1 GDAI01001116 JAI16487.1 KQ978287 KYM95609.1 GEBQ01022096 JAT17881.1 GL888103 EGI67352.1 GECU01030233 JAS77473.1 DS235327 EEB14726.1 LJIJ01000033 ODN04921.1 GALA01001615 JAA93237.1 BT126897 AEE61859.1 AXCN02002153 GFDF01003886 JAV10198.1 KB632256 ERL90675.1 APGK01050872 KB741178 ENN73159.1 CH477807 EAT36034.1 APCN01001262 AAAB01008846 EAA06504.5 CVRI01000044 CRK96466.1 GFXV01004487 MBW16292.1 UFQS01001175 UFQS01001201 UFQT01001175 UFQT01001201 SSX09411.1 SSX09695.1 SSX29313.1 GGFL01000882 MBW65060.1 GGFL01000883 MBW65061.1 GGFM01003668 MBW24419.1 CH916370 EDV99694.1 ATLV01014939 ATLV01014940 KE524996 KFB39667.1 JXJN01008099 GEDC01023789 GEDC01022599 GEDC01011803 GEDC01011376 GEDC01008019 JAS13509.1 JAS14699.1 JAS25495.1 JAS25922.1 JAS29279.1 GGMR01003384 MBY16003.1 GGFM01003674 MBW24425.1 GEDC01004656 JAS32642.1 ACPB03027316 GDKW01002279 JAI54316.1 ABLF02022575 KQ971372 EFA09453.1 GBGD01001969 JAC86920.1 LNIX01000006 OXA52810.1 AXCM01004618 CCAG010008024 EZ423130 ADD19406.1 GECL01002232 JAP03892.1 GGFK01005420 MBW38741.1 GBBI01000271 JAC18441.1 GAMD01000049 JAB01542.1 CM000162 EDX01324.1 KA646249 AFP60878.1 GGFJ01006367 MBW55508.1 CP012528 ALC48196.1 CH954180 EDV45681.1 CH379064 EDY72594.1 AE014298 AAS65258.1 AFH07223.1 AHN59305.1 AL133506 AY118486 LRGB01001579 KZS11574.1 CM000366 EDX16958.1 GBXI01016876 JAC97415.1 GDIP01224987 JAI98414.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000235965
UP000027135
+ More
UP000245037 UP000078540 UP000005205 UP000078492 UP000053825 UP000053105 UP000053097 UP000279307 UP000005203 UP000000311 UP000242457 UP000007266 UP000036403 UP000192223 UP000078542 UP000007755 UP000009046 UP000075884 UP000094527 UP000075886 UP000030742 UP000019118 UP000008820 UP000075902 UP000076407 UP000075903 UP000075840 UP000002358 UP000007062 UP000183832 UP000001070 UP000030765 UP000092460 UP000075920 UP000092443 UP000015103 UP000007819 UP000091820 UP000198287 UP000078200 UP000075883 UP000092444 UP000092445 UP000069272 UP000075900 UP000002282 UP000192221 UP000092553 UP000008711 UP000001819 UP000095301 UP000000803 UP000076858 UP000000304
UP000245037 UP000078540 UP000005205 UP000078492 UP000053825 UP000053105 UP000053097 UP000279307 UP000005203 UP000000311 UP000242457 UP000007266 UP000036403 UP000192223 UP000078542 UP000007755 UP000009046 UP000075884 UP000094527 UP000075886 UP000030742 UP000019118 UP000008820 UP000075902 UP000076407 UP000075903 UP000075840 UP000002358 UP000007062 UP000183832 UP000001070 UP000030765 UP000092460 UP000075920 UP000092443 UP000015103 UP000007819 UP000091820 UP000198287 UP000078200 UP000075883 UP000092444 UP000092445 UP000069272 UP000075900 UP000002282 UP000192221 UP000092553 UP000008711 UP000001819 UP000095301 UP000000803 UP000076858 UP000000304
Gene 3D
CDD
ProteinModelPortal
H9J7L2
A0A2H1VHE9
A0A194PQN4
A0A194QSQ3
S4Q095
A0A1E1WNR6
+ More
A0A2A4IYH0 A0A2A4IXW2 A0A2J7Q8J3 A0A067RTI7 A0A2P8YR06 A0A195AVG2 A0A158NF48 A0A195E8C6 A0A0L7R8M0 A0A0M8ZSJ5 E9ID26 A0A026W2R5 A0A088APV8 E2B080 A0A2A3EI99 D6WH93 A0A1Y1N0H1 A0A0J7KY31 A0A1Q3FYR5 A0A0T6BE85 A0A1W4X4K8 A0A023EQX9 T1DEC3 A0A0K8TQ19 A0A195C425 A0A1B6L2L4 F4WEI9 A0A1B6HS06 E0VMX0 A0A182N8F2 A0A1D2NI48 T1E212 J3JUI3 A0A182Q6N7 A0A1L8DUU0 U4UKY9 N6SZG0 Q16NV2 A0A182U4M5 A0A182XBX6 A0A182UVS2 A0A182IAK7 K7IRZ8 Q7QF35 A0A1J1I9Q1 A0A2H8TRS2 A0A336MKB9 A0A2M4CI84 A0A2M4CIU0 A0A2M3Z7B0 B4JJE2 A0A084VNX3 A0A1B0B3X0 A0A182W5L5 A0A1B6DIN6 A0A2S2NFL9 A0A1A9XVI2 A0A2M3Z7E4 A0A1B6E3Y5 T1HQX1 A0A0P4VJF3 J9JQL9 D6X2J7 A0A069DSM7 A0A1A9W971 A0A226E637 A0A1A9V0K9 A0A182M9S0 D3TNX8 A0A1B0A8G5 A0A0V0G760 A0A182FIR7 A0A2M4AD83 A0A023FAD9 A0A182RHK2 T1DUG4 B4Q0V7 A0A1W4VS75 T1PC33 A0A2M4BRU8 A0A0M4ELC3 B3NTX3 B5DM77 A0A1W4VFI4 A0A1I8MKA9 A4V3W8 Q9U1M2 A0A162DGT0 B4R3U6 A0A0A1WFG4 A0A0P4YN02
A0A2A4IYH0 A0A2A4IXW2 A0A2J7Q8J3 A0A067RTI7 A0A2P8YR06 A0A195AVG2 A0A158NF48 A0A195E8C6 A0A0L7R8M0 A0A0M8ZSJ5 E9ID26 A0A026W2R5 A0A088APV8 E2B080 A0A2A3EI99 D6WH93 A0A1Y1N0H1 A0A0J7KY31 A0A1Q3FYR5 A0A0T6BE85 A0A1W4X4K8 A0A023EQX9 T1DEC3 A0A0K8TQ19 A0A195C425 A0A1B6L2L4 F4WEI9 A0A1B6HS06 E0VMX0 A0A182N8F2 A0A1D2NI48 T1E212 J3JUI3 A0A182Q6N7 A0A1L8DUU0 U4UKY9 N6SZG0 Q16NV2 A0A182U4M5 A0A182XBX6 A0A182UVS2 A0A182IAK7 K7IRZ8 Q7QF35 A0A1J1I9Q1 A0A2H8TRS2 A0A336MKB9 A0A2M4CI84 A0A2M4CIU0 A0A2M3Z7B0 B4JJE2 A0A084VNX3 A0A1B0B3X0 A0A182W5L5 A0A1B6DIN6 A0A2S2NFL9 A0A1A9XVI2 A0A2M3Z7E4 A0A1B6E3Y5 T1HQX1 A0A0P4VJF3 J9JQL9 D6X2J7 A0A069DSM7 A0A1A9W971 A0A226E637 A0A1A9V0K9 A0A182M9S0 D3TNX8 A0A1B0A8G5 A0A0V0G760 A0A182FIR7 A0A2M4AD83 A0A023FAD9 A0A182RHK2 T1DUG4 B4Q0V7 A0A1W4VS75 T1PC33 A0A2M4BRU8 A0A0M4ELC3 B3NTX3 B5DM77 A0A1W4VFI4 A0A1I8MKA9 A4V3W8 Q9U1M2 A0A162DGT0 B4R3U6 A0A0A1WFG4 A0A0P4YN02
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
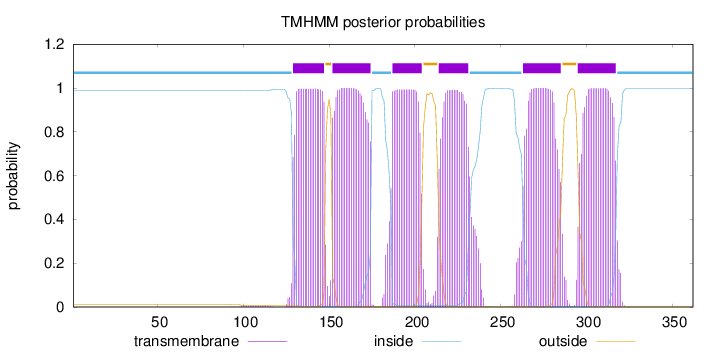
Length:
362
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
123.88878
Exp number, first 60 AAs:
0
Total prob of N-in:
0.98920
inside
1 - 128
TMhelix
129 - 147
outside
148 - 151
TMhelix
152 - 174
inside
175 - 186
TMhelix
187 - 204
outside
205 - 213
TMhelix
214 - 231
inside
232 - 262
TMhelix
263 - 285
outside
286 - 294
TMhelix
295 - 317
inside
318 - 362
Population Genetic Test Statistics
Pi
189.970302
Theta
170.702573
Tajima's D
0.578972
CLR
0.105861
CSRT
0.541422928853557
Interpretation
Uncertain
