Gene
KWMTBOMO04625 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005500
Annotation
PREDICTED:_beta-glucuronidase_[Bombyx_mori]
Full name
Beta-glucuronidase
Location in the cell
PlasmaMembrane Reliability : 1.477
Sequence
CDS
ATGATAAAGCCGAGATTTGCTTTCGCGCTTCTGCTAATATGCATAGTGTCCGTGACGATAGCAGTGGTGCCGCTGTCGTCTAGCTCAAATGAAATCAACCAATTGTATAAAAGAAAATTACCAAGCAACGGAGGCGCATTGTACCCACGTGCTACAGAAACCAGGGACTTAAGAACCCTAGATGGGATATGGAGTTTTCGGCCATCGCCTGCCGACCCAGAATTCGGCTATAGAAATGGCTGGTATGCGCAAGATCTTGAAAAGACAGGCTCTGTGATACACATGCCTGTGCCTTCTTCATATAATGACGTGGGCGAAGACGCCTCTCTTCGGGACCATGTAGGCTTGGTGTGGTACGACAGGAGGTTCCACGTTCCGCCGTGGTGGCAGACGGCGAAACAACGGGTCTGGGTGCGGTTCAGCAGCGTTCACTATGCCGCCGAAGTGTTTGTAAACGGCAAATCAGTGACATATCACGAGATAGGTCACTTACCGTTCGAAGTAGAGATCACGGATGTAGTGAATTACAATACGAGCAATCTCCTCACTGTTGCGGTCGACAACACCCTACTAAGCGATACTGTACCACAGGGATATGTCAAACAAATTATCGATTTTGATAAAGTGCGACAGGAGCAAACATACACGTTTGATTTTTTCAATTACGCTGGTATTCATAGGACTGTATTCTTATATTCAACGCCCCAAGTTTATATTGACGATGTTATTGTTAACACCGACATTCAAGGATTCACAGGATTCGTGGTATACAACGTAACTCACAAGGGGCCGACCACAGAAGACGTTCTCTGCCTCATTCAAGTGTTCGATAAGTCGGACACACAAGTCGTGGCCTCAAACGAGTGCGCCGGCCTATTGGAAATAGCGAATGCTAGATTCTGGTGGCCTTACCTCATGAACCCCGAGCCGGGATACTTGTACACTTTTAGGGCTTCTCTAATAAGTAGACGCGGAGATATAATCGACACTTACAGTCAGAAGATCGGTATTAGAACGGTCACGTGGACAAATACAACCATATACTTGAATTCGAAACCATTATATCTGAGGGGATTTGGGATGCACGAAGACTCGGATCTCAGAGGCAAAGGATGGGACCCAGTTTTGTGGGTGAAGAATTTTAACTTGATCAAATGGATCGGAGCGAACGCGTTCCGGACATCGCATTATCCGTACGCTGAGGAAATATACCAGTTGGCCGACGAATATGGGATAATGATCATTGATGAGTGTCCCAGCGGAGACACAGACATCTTCACGCAGTCTCTACTGAATAAGCACAAACAATCACTGACGGAGCTCATCAGACGCGACAAGAATCACCCGAGCGTCATCATGTGGTCCATCTCCAACGAGCCGCGGTCTGCCAAACAACAGGCCGATGCCTATTTCGCTGAAGTAGTGAAACACGTGAAGTCCATGGACCTCTCCAGACCCGTCACCATCGCGATCTCGCAATATTACAACACTGATAGAGCAGGACAACACTTAGACGTCATCTGCTTCAACCGTTACAACGGCTGGTACCAAAACACCGGCGACTTGAACTACATCGTCAGTAAGGTGGTGGAAGAAGCCACCGAGTTCCACCGGCGACACAACAAGCCGGTCATCATGACCGAATATGGCGCTGACACTGTTGCCGGCCTACACTTTATTCCAGAATATGTGTGGTCTGAAGAATACCAAGTGGCGCTGATGTCAGAACATTTCAAAGCGTTCGATAAACTGAGACAGACCGGCTTCTTCGCTGGCGAGTTCATATGGAATTTCGCGGATTTCAAAACAGCGCAAACGATAACCCGCGTCGGCGGCAACAGGAAAGGTATATTCACTCGGTCGCGGCAGCCTAAAGCGTCCGCGCACCACCTCCGCACGCGCTATCTGTCGCTCGCTGTCGCCGACGGCGCCGCCAATATACCGGACCCAACGTACTACATCTCCGACAACAAGCCCACCCTACACGAAGATCTATAA
Protein
MIKPRFAFALLLICIVSVTIAVVPLSSSSNEINQLYKRKLPSNGGALYPRATETRDLRTLDGIWSFRPSPADPEFGYRNGWYAQDLEKTGSVIHMPVPSSYNDVGEDASLRDHVGLVWYDRRFHVPPWWQTAKQRVWVRFSSVHYAAEVFVNGKSVTYHEIGHLPFEVEITDVVNYNTSNLLTVAVDNTLLSDTVPQGYVKQIIDFDKVRQEQTYTFDFFNYAGIHRTVFLYSTPQVYIDDVIVNTDIQGFTGFVVYNVTHKGPTTEDVLCLIQVFDKSDTQVVASNECAGLLEIANARFWWPYLMNPEPGYLYTFRASLISRRGDIIDTYSQKIGIRTVTWTNTTIYLNSKPLYLRGFGMHEDSDLRGKGWDPVLWVKNFNLIKWIGANAFRTSHYPYAEEIYQLADEYGIMIIDECPSGDTDIFTQSLLNKHKQSLTELIRRDKNHPSVIMWSISNEPRSAKQQADAYFAEVVKHVKSMDLSRPVTIAISQYYNTDRAGQHLDVICFNRYNGWYQNTGDLNYIVSKVVEEATEFHRRHNKPVIMTEYGADTVAGLHFIPEYVWSEEYQVALMSEHFKAFDKLRQTGFFAGEFIWNFADFKTAQTITRVGGNRKGIFTRSRQPKASAHHLRTRYLSLAVADGAANIPDPTYYISDNKPTLHEDL
Summary
Description
Plays an important role in the degradation of dermatan and keratan sulfates.
Catalytic Activity
a beta-D-glucuronoside + H2O = an alcohol + D-glucuronate
Subunit
Homotetramer.
Similarity
Belongs to the glycosyl hydrolase 2 family.
Feature
chain Beta-glucuronidase
Uniprot
H9J7K8
A0A212FK42
A0A194QSP8
A0A194PRN5
A0A2H1VVM2
A0A2J7QP48
+ More
A0A067QW61 A0A1W4WJ63 A0A1Y1NAJ6 A0A084WIR1 D6WYY8 A0A1S4G6Y4 A0A0P4VLU9 A0A182QNR6 A0A1B0CV53 J9HEW9 A0A1I8M8H2 A0A182RRY1 A0A1L8E1S4 A0A1L8E203 A0A023EVQ3 A0A1L8E1Q0 A0A1I8M8I1 T1HP51 B4KU73 A0A0Q9X8T4 A0A2H8TWI1 A0A1J1I8L2 A0A0V0G5X7 N6TPQ9 A0A0A1WSE6 A0A182N3Z2 A0A1J1I4S8 A0A0A1XQ59 J9K812 A0A0A1X7P8 A0A0Q9XB98 A0A182JHB8 A0A0R3NQ73 A0A0P8XQ24 A0A3B0JJC2 A0A3B0JNB6 A0A2S2QLY1 A0A3B0J1I2 A0A3B0JH71 A0A3B0IY96 A0A0R3NVC0 Q28WJ8 A0A182WDW6 A0A182WDW7 A0A182WDW8 A0A3B0JBP6 A0A0R3NWJ6 B4H723 B3MCH6 A0A182WDW5 A0A0P8XQ09 A0A0R3NQG8 A0A182XGU9 A0A0P9AFR9 A0A182L502 W5JQR0 A0A0Q9XG25 A0A034WSD2 W8B1E3 A0A182V7U1 A0A0Q5VYG2 A0A0K8VYY1 A0A034WPW2 B3NKF1 A0A182HHG9 A0A1Q3FG12 W8BNG7 D6WYY7 A0A0M5IZS1 A0A182MGY9 A0A1Q3FG15 A0A1W4V6D4 A0A1W4VIQ9 A0A1I8PMS8 A0A1I8PMW3 W8B8D4 A0A1W4VJI7 A0A0K8SX62 A0A1I8PMR1 A0A182PUA5 E2AGJ7 A0A1I8PMW5 A0A0J9U5U1 A0A0J9RG28 A0A0A9XSU5 A0A182Y9D5 A0A0J9RGD8 A0A0J9RGD0 B4QDL7 C8VV29 A0A0A9Z159 A0A0Q9WVL5 A0A0K8SMX2
A0A067QW61 A0A1W4WJ63 A0A1Y1NAJ6 A0A084WIR1 D6WYY8 A0A1S4G6Y4 A0A0P4VLU9 A0A182QNR6 A0A1B0CV53 J9HEW9 A0A1I8M8H2 A0A182RRY1 A0A1L8E1S4 A0A1L8E203 A0A023EVQ3 A0A1L8E1Q0 A0A1I8M8I1 T1HP51 B4KU73 A0A0Q9X8T4 A0A2H8TWI1 A0A1J1I8L2 A0A0V0G5X7 N6TPQ9 A0A0A1WSE6 A0A182N3Z2 A0A1J1I4S8 A0A0A1XQ59 J9K812 A0A0A1X7P8 A0A0Q9XB98 A0A182JHB8 A0A0R3NQ73 A0A0P8XQ24 A0A3B0JJC2 A0A3B0JNB6 A0A2S2QLY1 A0A3B0J1I2 A0A3B0JH71 A0A3B0IY96 A0A0R3NVC0 Q28WJ8 A0A182WDW6 A0A182WDW7 A0A182WDW8 A0A3B0JBP6 A0A0R3NWJ6 B4H723 B3MCH6 A0A182WDW5 A0A0P8XQ09 A0A0R3NQG8 A0A182XGU9 A0A0P9AFR9 A0A182L502 W5JQR0 A0A0Q9XG25 A0A034WSD2 W8B1E3 A0A182V7U1 A0A0Q5VYG2 A0A0K8VYY1 A0A034WPW2 B3NKF1 A0A182HHG9 A0A1Q3FG12 W8BNG7 D6WYY7 A0A0M5IZS1 A0A182MGY9 A0A1Q3FG15 A0A1W4V6D4 A0A1W4VIQ9 A0A1I8PMS8 A0A1I8PMW3 W8B8D4 A0A1W4VJI7 A0A0K8SX62 A0A1I8PMR1 A0A182PUA5 E2AGJ7 A0A1I8PMW5 A0A0J9U5U1 A0A0J9RG28 A0A0A9XSU5 A0A182Y9D5 A0A0J9RGD8 A0A0J9RGD0 B4QDL7 C8VV29 A0A0A9Z159 A0A0Q9WVL5 A0A0K8SMX2
EC Number
3.2.1.31
Pubmed
EMBL
BABH01013989
AGBW02008170
OWR54070.1
KQ461154
KPJ08543.1
KQ459595
+ More
KPI95613.1 ODYU01004702 SOQ44863.1 NEVH01012097 PNF30365.1 KK853297 KDR08779.1 GEZM01013355 GEZM01013354 JAV92627.1 ATLV01023944 KE525347 KFB50105.1 KQ971363 EFA07841.1 GDKW01001145 JAI55450.1 AXCN02001092 AJWK01030100 CH477201 EJY57347.1 GFDF01001426 JAV12658.1 GFDF01001425 JAV12659.1 JXUM01129069 GAPW01000441 KQ567397 JAC13157.1 KXJ69450.1 GFDF01001424 JAV12660.1 ACPB03008079 CH933808 EDW09669.2 KRG04843.1 GFXV01006802 MBW18607.1 CVRI01000041 CRK95286.1 GECL01002723 JAP03401.1 APGK01056970 KB741277 ENN71225.1 GBXI01012530 JAD01762.1 CRK95285.1 GBXI01001136 JAD13156.1 ABLF02036573 GBXI01007584 JAD06708.1 KRG04841.1 CM000071 KRT03297.1 CH902619 KPU76703.1 OUUW01000001 SPP72701.1 SPP72698.1 GGMS01009562 MBY78765.1 SPP72702.1 SPP72699.1 SPP72697.1 KRT03295.1 EAL26669.3 SPP72700.1 KRT03296.1 CH479216 EDW33659.1 EDV37300.1 KPU76701.1 KRT03298.1 KPU76702.1 ADMH02000720 ETN65250.1 KRG04842.1 GAKP01002284 JAC56668.1 GAMC01011630 JAB94925.1 CH954179 KQS62122.1 GDHF01008544 JAI43770.1 GAKP01002283 JAC56669.1 EDV55173.1 APCN01002410 GFDL01008562 JAV26483.1 GAMC01011629 JAB94926.1 EFA07842.1 CP012524 ALC42124.1 AXCM01008037 GFDL01008547 JAV26498.1 GAMC01011631 GAMC01011628 GAMC01011627 JAB94927.1 GBRD01008070 JAG57751.1 GL439316 EFN67454.1 CM002911 KMY94985.1 KMY94983.1 GBHO01020565 JAG23039.1 KMY94986.1 KMY94982.1 CM000362 EDX07773.1 KMY94981.1 BT099775 ACV88434.1 GBHO01004593 GBHO01004592 JAG39011.1 JAG39012.1 CH940659 KRF85729.1 GBRD01011360 JAG54464.1
KPI95613.1 ODYU01004702 SOQ44863.1 NEVH01012097 PNF30365.1 KK853297 KDR08779.1 GEZM01013355 GEZM01013354 JAV92627.1 ATLV01023944 KE525347 KFB50105.1 KQ971363 EFA07841.1 GDKW01001145 JAI55450.1 AXCN02001092 AJWK01030100 CH477201 EJY57347.1 GFDF01001426 JAV12658.1 GFDF01001425 JAV12659.1 JXUM01129069 GAPW01000441 KQ567397 JAC13157.1 KXJ69450.1 GFDF01001424 JAV12660.1 ACPB03008079 CH933808 EDW09669.2 KRG04843.1 GFXV01006802 MBW18607.1 CVRI01000041 CRK95286.1 GECL01002723 JAP03401.1 APGK01056970 KB741277 ENN71225.1 GBXI01012530 JAD01762.1 CRK95285.1 GBXI01001136 JAD13156.1 ABLF02036573 GBXI01007584 JAD06708.1 KRG04841.1 CM000071 KRT03297.1 CH902619 KPU76703.1 OUUW01000001 SPP72701.1 SPP72698.1 GGMS01009562 MBY78765.1 SPP72702.1 SPP72699.1 SPP72697.1 KRT03295.1 EAL26669.3 SPP72700.1 KRT03296.1 CH479216 EDW33659.1 EDV37300.1 KPU76701.1 KRT03298.1 KPU76702.1 ADMH02000720 ETN65250.1 KRG04842.1 GAKP01002284 JAC56668.1 GAMC01011630 JAB94925.1 CH954179 KQS62122.1 GDHF01008544 JAI43770.1 GAKP01002283 JAC56669.1 EDV55173.1 APCN01002410 GFDL01008562 JAV26483.1 GAMC01011629 JAB94926.1 EFA07842.1 CP012524 ALC42124.1 AXCM01008037 GFDL01008547 JAV26498.1 GAMC01011631 GAMC01011628 GAMC01011627 JAB94927.1 GBRD01008070 JAG57751.1 GL439316 EFN67454.1 CM002911 KMY94985.1 KMY94983.1 GBHO01020565 JAG23039.1 KMY94986.1 KMY94982.1 CM000362 EDX07773.1 KMY94981.1 BT099775 ACV88434.1 GBHO01004593 GBHO01004592 JAG39011.1 JAG39012.1 CH940659 KRF85729.1 GBRD01011360 JAG54464.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000235965
UP000027135
+ More
UP000192223 UP000030765 UP000007266 UP000075886 UP000092461 UP000008820 UP000095301 UP000075900 UP000069940 UP000249989 UP000015103 UP000009192 UP000183832 UP000019118 UP000075884 UP000007819 UP000075880 UP000001819 UP000007801 UP000268350 UP000075920 UP000008744 UP000076407 UP000075882 UP000000673 UP000075903 UP000008711 UP000075840 UP000092553 UP000075883 UP000192221 UP000095300 UP000075885 UP000000311 UP000076408 UP000000304 UP000008792
UP000192223 UP000030765 UP000007266 UP000075886 UP000092461 UP000008820 UP000095301 UP000075900 UP000069940 UP000249989 UP000015103 UP000009192 UP000183832 UP000019118 UP000075884 UP000007819 UP000075880 UP000001819 UP000007801 UP000268350 UP000075920 UP000008744 UP000076407 UP000075882 UP000000673 UP000075903 UP000008711 UP000075840 UP000092553 UP000075883 UP000192221 UP000095300 UP000075885 UP000000311 UP000076408 UP000000304 UP000008792
Interpro
Gene 3D
ProteinModelPortal
H9J7K8
A0A212FK42
A0A194QSP8
A0A194PRN5
A0A2H1VVM2
A0A2J7QP48
+ More
A0A067QW61 A0A1W4WJ63 A0A1Y1NAJ6 A0A084WIR1 D6WYY8 A0A1S4G6Y4 A0A0P4VLU9 A0A182QNR6 A0A1B0CV53 J9HEW9 A0A1I8M8H2 A0A182RRY1 A0A1L8E1S4 A0A1L8E203 A0A023EVQ3 A0A1L8E1Q0 A0A1I8M8I1 T1HP51 B4KU73 A0A0Q9X8T4 A0A2H8TWI1 A0A1J1I8L2 A0A0V0G5X7 N6TPQ9 A0A0A1WSE6 A0A182N3Z2 A0A1J1I4S8 A0A0A1XQ59 J9K812 A0A0A1X7P8 A0A0Q9XB98 A0A182JHB8 A0A0R3NQ73 A0A0P8XQ24 A0A3B0JJC2 A0A3B0JNB6 A0A2S2QLY1 A0A3B0J1I2 A0A3B0JH71 A0A3B0IY96 A0A0R3NVC0 Q28WJ8 A0A182WDW6 A0A182WDW7 A0A182WDW8 A0A3B0JBP6 A0A0R3NWJ6 B4H723 B3MCH6 A0A182WDW5 A0A0P8XQ09 A0A0R3NQG8 A0A182XGU9 A0A0P9AFR9 A0A182L502 W5JQR0 A0A0Q9XG25 A0A034WSD2 W8B1E3 A0A182V7U1 A0A0Q5VYG2 A0A0K8VYY1 A0A034WPW2 B3NKF1 A0A182HHG9 A0A1Q3FG12 W8BNG7 D6WYY7 A0A0M5IZS1 A0A182MGY9 A0A1Q3FG15 A0A1W4V6D4 A0A1W4VIQ9 A0A1I8PMS8 A0A1I8PMW3 W8B8D4 A0A1W4VJI7 A0A0K8SX62 A0A1I8PMR1 A0A182PUA5 E2AGJ7 A0A1I8PMW5 A0A0J9U5U1 A0A0J9RG28 A0A0A9XSU5 A0A182Y9D5 A0A0J9RGD8 A0A0J9RGD0 B4QDL7 C8VV29 A0A0A9Z159 A0A0Q9WVL5 A0A0K8SMX2
A0A067QW61 A0A1W4WJ63 A0A1Y1NAJ6 A0A084WIR1 D6WYY8 A0A1S4G6Y4 A0A0P4VLU9 A0A182QNR6 A0A1B0CV53 J9HEW9 A0A1I8M8H2 A0A182RRY1 A0A1L8E1S4 A0A1L8E203 A0A023EVQ3 A0A1L8E1Q0 A0A1I8M8I1 T1HP51 B4KU73 A0A0Q9X8T4 A0A2H8TWI1 A0A1J1I8L2 A0A0V0G5X7 N6TPQ9 A0A0A1WSE6 A0A182N3Z2 A0A1J1I4S8 A0A0A1XQ59 J9K812 A0A0A1X7P8 A0A0Q9XB98 A0A182JHB8 A0A0R3NQ73 A0A0P8XQ24 A0A3B0JJC2 A0A3B0JNB6 A0A2S2QLY1 A0A3B0J1I2 A0A3B0JH71 A0A3B0IY96 A0A0R3NVC0 Q28WJ8 A0A182WDW6 A0A182WDW7 A0A182WDW8 A0A3B0JBP6 A0A0R3NWJ6 B4H723 B3MCH6 A0A182WDW5 A0A0P8XQ09 A0A0R3NQG8 A0A182XGU9 A0A0P9AFR9 A0A182L502 W5JQR0 A0A0Q9XG25 A0A034WSD2 W8B1E3 A0A182V7U1 A0A0Q5VYG2 A0A0K8VYY1 A0A034WPW2 B3NKF1 A0A182HHG9 A0A1Q3FG12 W8BNG7 D6WYY7 A0A0M5IZS1 A0A182MGY9 A0A1Q3FG15 A0A1W4V6D4 A0A1W4VIQ9 A0A1I8PMS8 A0A1I8PMW3 W8B8D4 A0A1W4VJI7 A0A0K8SX62 A0A1I8PMR1 A0A182PUA5 E2AGJ7 A0A1I8PMW5 A0A0J9U5U1 A0A0J9RG28 A0A0A9XSU5 A0A182Y9D5 A0A0J9RGD8 A0A0J9RGD0 B4QDL7 C8VV29 A0A0A9Z159 A0A0Q9WVL5 A0A0K8SMX2
PDB
3HN3
E-value=2.73564e-146,
Score=1332
Ontologies
PATHWAY
00040
Pentose and glucuronate interconversions - Bombyx mori (domestic silkworm)
00531 Glycosaminoglycan degradation - Bombyx mori (domestic silkworm)
00860 Porphyrin and chlorophyll metabolism - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
04142 Lysosome - Bombyx mori (domestic silkworm)
00531 Glycosaminoglycan degradation - Bombyx mori (domestic silkworm)
00860 Porphyrin and chlorophyll metabolism - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
04142 Lysosome - Bombyx mori (domestic silkworm)
GO
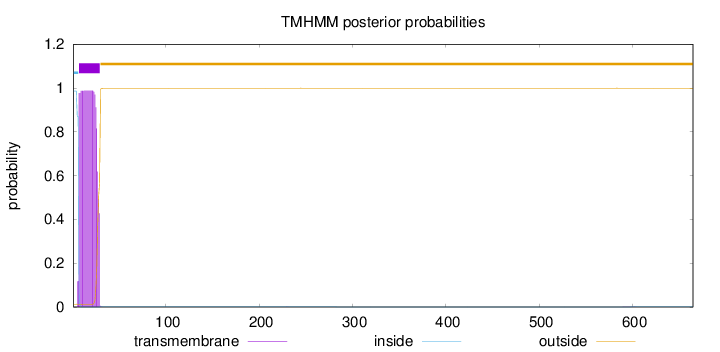
Topology
SignalP
Position: 1 - 21,
Likelihood: 0.872973
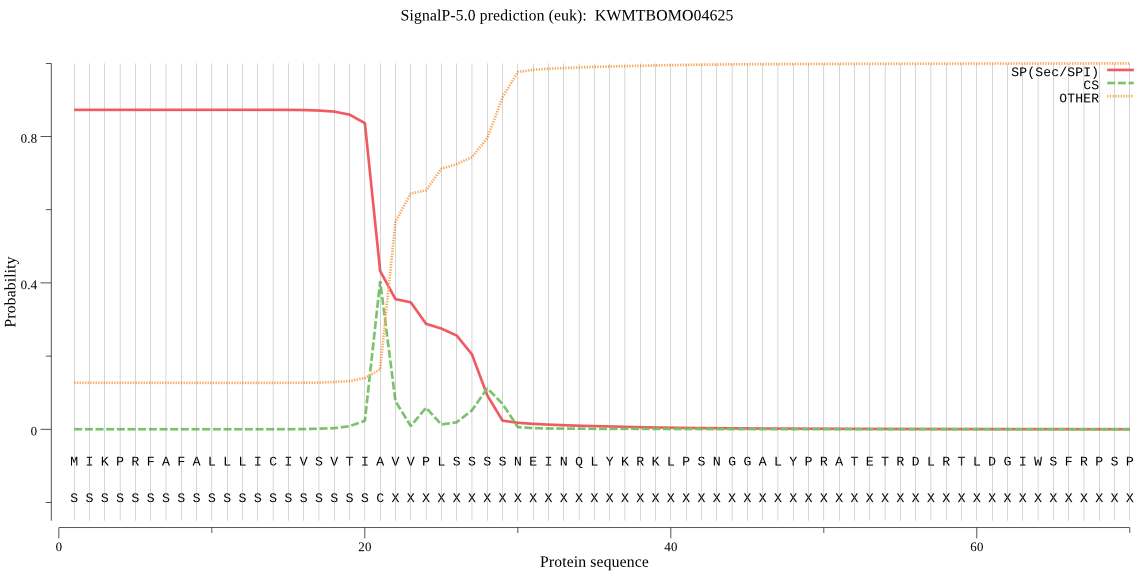
Length:
665
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.2527899999999
Exp number, first 60 AAs:
21.24357
Total prob of N-in:
0.98703
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 665
Population Genetic Test Statistics
Pi
202.409412
Theta
150.996145
Tajima's D
0.669917
CLR
0.738249
CSRT
0.565471726413679
Interpretation
Uncertain
