Gene
KWMTBOMO04624
Annotation
Matrix_metalloproteinase-16_[Papilio_machaon]
Full name
Matrix metalloproteinase-2
Location in the cell
Nuclear Reliability : 2.757
Sequence
CDS
ATGGACGGAAACGAGCAATCTTACGAAGACGATTTCCGGATCGCGATCAAAACTTTACAAGAATTCGGAGGTATTCCTGTCACAGGGGAGCTAGACGCAGCGACAAGAAATTTAATGAAACAAAAGCGGTGTGGTAGACCAGACAGAGAAGAACAGGATCACGAAAATCGAAGATCGAAGAGATTTGCCGTCCAGGGAGCGAAGTGGAAACACACCAATTTAACATGGAGGTAA
Protein
MDGNEQSYEDDFRIAIKTLQEFGGIPVTGELDAATRNLMKQKRCGRPDREEQDHENRRSKRFAVQGAKWKHTNLTWR
Summary
Description
Has metalloproteinase activity (PubMed:11967260). Required for larval tissue histolysis during metamorphosis and is involved in pupal head eversion and fusion of the wing imaginal tissue (PubMed:12530966). Required for growth of the dorsal air sac primordium and development of the dorsal air sacs (PubMed:19751719). Promotes embryonic motor axon fasciculation (PubMed:18045838). Cleaves and activates frac to promote motor axon bundling during outgrowth (PubMed:21471368). Promotes the reshaping of adult sensory neuron dendrites from a radial to lattice-like shape which occurs after eclosion by degrading the basement membrane on which the dendrites grow (PubMed:20412776). Involved in inhibition of follicle stem cell proliferation by cleaving Dlp, inhibiting its interaction with wg and preventing Dlp-mediated spreading of wg to follicle stem cells to enhance their proliferation (PubMed:25267296). Plays a role in wound healing (PubMed:22262460). Involved in fat body dissociation which occurs during metamorphosis by degrading basement membrane components, leading to destruction of cell-basement membrane junctions (PubMed:25520167). Required for posterior follicle cell degradation and ovulation (PubMed:25695427).
Cofactor
Zn(2+)
Ca(2+)
Ca(2+)
Similarity
Belongs to the peptidase M10A family.
Keywords
Alternative splicing
Calcium
Cell membrane
Cleavage on pair of basic residues
Complete proteome
Disulfide bond
Hydrolase
Membrane
Metal-binding
Metalloprotease
Protease
Reference proteome
Repeat
Signal
Transmembrane
Transmembrane helix
Zinc
Zymogen
Feature
propeptide Activation peptide
chain Matrix metalloproteinase-2
splice variant In isoform C.
chain Matrix metalloproteinase-2
splice variant In isoform C.
Uniprot
A0A2A4IWM9
A0A2H1VB64
A0A194QU76
A0A194PQM8
A0A212FGS1
A0A0L7LT68
+ More
A0A336N0B2 A0A0J7L3C7 E9IWG0 A0A336MJP8 E2BGY0 E2AGJ9 A0A195FKY6 A0A087ZQ14 A0A1Y1LH94 A0A2A3EGV3 T1KNP2 A0A2S2NLZ5 A0A151IPY0 A0A0P5H5I2 A0A1A9V8N5 A0A183IHM5 A0A2H8TRK4 A0A158NET9 A0A195AV09 A0A348G5Y9 A0A232FL65 A0A151XIP8 A0A1B0G123 A0A3L8DAM3 A0A1B0ARG3 A0A195E8Z7 A0A1B0AD89 A0A1A9W7U8 A0A1J1I9A7 A0A0M3HUN9 A0A1B0GI40 A0A1A9YH87 A0A1S3DAR8 A0A0T6B4Y1 A0A164KHU5 A0A0L7R185 H2ZEY6 A0A1B6E667 D6WYY9 A0A147BPY1 A0A2S2Q4U3 T2M4Y7 A0A1I8MCL0 A0A0P4ZTH9 A0A0P5BVX1 A0A0P6BPJ7 X1WIM9 A0A1W4WJ61 B7QD72 B4HT26 A0A0P6IFT8 A0A0P6F6M4 F6RRU7 A0A0S3Q290 A0A1B6G040 A0A087USP2 A0A1W2W5Z1 A0A3M6V0B5 E0V8V4 A0A0V1N7S9 A0A0V1HWH9 Q8MPP3 E0VM71 A0A0L0BR13 A0A0V1EZ73 A0A0V1HWB5 A0A0V1JCY4 A0A0V0YC80 A0A0N5D346 A0A0V1EYX9 A0A0V1JD56 A0A1B0EX88 A0A1S3IGL5 T2M2R4 B4QHP4 Q9U9P0 E9GW38 H2ZEY7 A0A3P6UHJ2 A0A1W0XBN1 A0A3B3E0A3 A0A3B0JKN2 A0A0J9RA38 A0A1D1VJZ5 A0A0J9R8Z6 A0A1I7VM22 A0A1B0GPG4 A0A1B6HAB5 A0A2P6L761 A0A3Q0HIT9 A0A0C9QWS2 A0A0V0SA94 W8AXQ9 A0A2P2I1R8
A0A336N0B2 A0A0J7L3C7 E9IWG0 A0A336MJP8 E2BGY0 E2AGJ9 A0A195FKY6 A0A087ZQ14 A0A1Y1LH94 A0A2A3EGV3 T1KNP2 A0A2S2NLZ5 A0A151IPY0 A0A0P5H5I2 A0A1A9V8N5 A0A183IHM5 A0A2H8TRK4 A0A158NET9 A0A195AV09 A0A348G5Y9 A0A232FL65 A0A151XIP8 A0A1B0G123 A0A3L8DAM3 A0A1B0ARG3 A0A195E8Z7 A0A1B0AD89 A0A1A9W7U8 A0A1J1I9A7 A0A0M3HUN9 A0A1B0GI40 A0A1A9YH87 A0A1S3DAR8 A0A0T6B4Y1 A0A164KHU5 A0A0L7R185 H2ZEY6 A0A1B6E667 D6WYY9 A0A147BPY1 A0A2S2Q4U3 T2M4Y7 A0A1I8MCL0 A0A0P4ZTH9 A0A0P5BVX1 A0A0P6BPJ7 X1WIM9 A0A1W4WJ61 B7QD72 B4HT26 A0A0P6IFT8 A0A0P6F6M4 F6RRU7 A0A0S3Q290 A0A1B6G040 A0A087USP2 A0A1W2W5Z1 A0A3M6V0B5 E0V8V4 A0A0V1N7S9 A0A0V1HWH9 Q8MPP3 E0VM71 A0A0L0BR13 A0A0V1EZ73 A0A0V1HWB5 A0A0V1JCY4 A0A0V0YC80 A0A0N5D346 A0A0V1EYX9 A0A0V1JD56 A0A1B0EX88 A0A1S3IGL5 T2M2R4 B4QHP4 Q9U9P0 E9GW38 H2ZEY7 A0A3P6UHJ2 A0A1W0XBN1 A0A3B3E0A3 A0A3B0JKN2 A0A0J9RA38 A0A1D1VJZ5 A0A0J9R8Z6 A0A1I7VM22 A0A1B0GPG4 A0A1B6HAB5 A0A2P6L761 A0A3Q0HIT9 A0A0C9QWS2 A0A0V0SA94 W8AXQ9 A0A2P2I1R8
EC Number
3.4.24.-
Pubmed
26354079
22118469
26227816
21282665
20798317
28004739
+ More
21347285 28648823 30249741 18362917 19820115 29652888 24065732 25315136 17994087 12481130 15114417 30382153 20566863 11967260 10731132 12537572 12530966 18045838 19751719 20412776 21471368 22262460 25267296 25520167 25695427 26108605 10648248 21292972 29451363 22936249 27649274 25217238 24495485
21347285 28648823 30249741 18362917 19820115 29652888 24065732 25315136 17994087 12481130 15114417 30382153 20566863 11967260 10731132 12537572 12530966 18045838 19751719 20412776 21471368 22262460 25267296 25520167 25695427 26108605 10648248 21292972 29451363 22936249 27649274 25217238 24495485
EMBL
NWSH01006225
PCG63582.1
ODYU01001602
SOQ38051.1
KQ461154
KPJ08540.1
+ More
KQ459595 KPI95612.1 AGBW02008605 OWR52927.1 JTDY01000150 KOB78584.1 UFQS01005597 UFQT01005597 SSX16855.1 SSX36032.1 LBMM01001098 KMQ96929.1 GL766526 EFZ15070.1 UFQT01001129 SSX29063.1 GL448228 EFN85112.1 GL439316 EFN67456.1 KQ981490 KYN41330.1 GEZM01055573 JAV73019.1 KZ288254 PBC30704.1 CAEY01000277 GGMR01005590 MBY18209.1 KQ976806 KYN08207.1 GDIQ01254645 JAJ97079.1 UZAM01007580 VDP00079.1 GFXV01005022 MBW16827.1 ADTU01013670 ADTU01013671 ADTU01013672 ADTU01013673 ADTU01013674 ADTU01013675 ADTU01013676 ADTU01013677 ADTU01013678 ADTU01013679 KQ976736 KYM76078.1 FX985527 BBF97862.1 NNAY01000055 OXU31481.1 KQ982080 KYQ60201.1 CCAG010005036 QOIP01000010 RLU17361.1 JXJN01002410 KQ979479 KYN21302.1 CVRI01000044 CRK96840.1 AJWK01011259 AJWK01011260 LJIG01009817 KRT82321.1 LRGB01003325 KZS03275.1 KQ414668 KOC64604.1 GEDC01025185 GEDC01003893 JAS12113.1 JAS33405.1 KQ971363 EFA09264.2 GEGO01002587 JAR92817.1 GGMS01003418 MBY72621.1 HAAD01000947 CDG67179.1 GDIP01208779 JAJ14623.1 GDIP01179180 JAJ44222.1 GDIP01012120 JAM91595.1 ABLF02036564 ABLF02036569 ABLF02036574 ABJB010380367 ABJB010586123 DS911299 EEC16794.1 CH480816 EDW47136.1 GDIQ01005176 JAN89561.1 GDIQ01064753 JAN29984.1 EAAA01000357 LC056002 BAT62454.1 GECZ01014096 JAS55673.1 KK121385 KFM80381.1 RCHS01000379 RMX59317.1 DS234986 EEB09810.1 JYDO01000004 KRZ80010.1 JYDP01000022 KRZ14720.1 AJ289232 AE013599 BT021212 BT150239 AAX33360.1 AGV77141.1 CAC81969.1 DS235291 EEB14477.1 JRES01001487 KNC22520.1 JYDR01000003 KRY78876.1 KRZ14721.1 JYDS01000013 JYDV01000001 KRZ32835.1 KRZ46247.1 JYDU01000025 KRX98046.1 UYYF01004496 VDN04767.1 KRY78875.1 KRZ32836.1 AJWK01011251 AJWK01011252 AJWK01011253 AJWK01011254 HAAD01000070 CDG66302.1 CM000362 EDX06378.1 AF162688 AAD45804.1 GL732569 EFX76334.1 UYRX01000250 VDK78558.1 MTYJ01000005 OQV24830.1 OUUW01000001 SPP74029.1 CM002911 KMY92554.1 BDGG01000005 GAU98818.1 KMY92553.1 JH712088 EJD76154.1 AJVK01015207 AJVK01015208 AJVK01015209 GECU01036129 JAS71577.1 MWRG01001283 PRD34414.1 GBYB01005122 JAG74889.1 JYDL01000025 KRX23301.1 GAMC01016967 JAB89588.1 IACF01002329 LAB67987.1
KQ459595 KPI95612.1 AGBW02008605 OWR52927.1 JTDY01000150 KOB78584.1 UFQS01005597 UFQT01005597 SSX16855.1 SSX36032.1 LBMM01001098 KMQ96929.1 GL766526 EFZ15070.1 UFQT01001129 SSX29063.1 GL448228 EFN85112.1 GL439316 EFN67456.1 KQ981490 KYN41330.1 GEZM01055573 JAV73019.1 KZ288254 PBC30704.1 CAEY01000277 GGMR01005590 MBY18209.1 KQ976806 KYN08207.1 GDIQ01254645 JAJ97079.1 UZAM01007580 VDP00079.1 GFXV01005022 MBW16827.1 ADTU01013670 ADTU01013671 ADTU01013672 ADTU01013673 ADTU01013674 ADTU01013675 ADTU01013676 ADTU01013677 ADTU01013678 ADTU01013679 KQ976736 KYM76078.1 FX985527 BBF97862.1 NNAY01000055 OXU31481.1 KQ982080 KYQ60201.1 CCAG010005036 QOIP01000010 RLU17361.1 JXJN01002410 KQ979479 KYN21302.1 CVRI01000044 CRK96840.1 AJWK01011259 AJWK01011260 LJIG01009817 KRT82321.1 LRGB01003325 KZS03275.1 KQ414668 KOC64604.1 GEDC01025185 GEDC01003893 JAS12113.1 JAS33405.1 KQ971363 EFA09264.2 GEGO01002587 JAR92817.1 GGMS01003418 MBY72621.1 HAAD01000947 CDG67179.1 GDIP01208779 JAJ14623.1 GDIP01179180 JAJ44222.1 GDIP01012120 JAM91595.1 ABLF02036564 ABLF02036569 ABLF02036574 ABJB010380367 ABJB010586123 DS911299 EEC16794.1 CH480816 EDW47136.1 GDIQ01005176 JAN89561.1 GDIQ01064753 JAN29984.1 EAAA01000357 LC056002 BAT62454.1 GECZ01014096 JAS55673.1 KK121385 KFM80381.1 RCHS01000379 RMX59317.1 DS234986 EEB09810.1 JYDO01000004 KRZ80010.1 JYDP01000022 KRZ14720.1 AJ289232 AE013599 BT021212 BT150239 AAX33360.1 AGV77141.1 CAC81969.1 DS235291 EEB14477.1 JRES01001487 KNC22520.1 JYDR01000003 KRY78876.1 KRZ14721.1 JYDS01000013 JYDV01000001 KRZ32835.1 KRZ46247.1 JYDU01000025 KRX98046.1 UYYF01004496 VDN04767.1 KRY78875.1 KRZ32836.1 AJWK01011251 AJWK01011252 AJWK01011253 AJWK01011254 HAAD01000070 CDG66302.1 CM000362 EDX06378.1 AF162688 AAD45804.1 GL732569 EFX76334.1 UYRX01000250 VDK78558.1 MTYJ01000005 OQV24830.1 OUUW01000001 SPP74029.1 CM002911 KMY92554.1 BDGG01000005 GAU98818.1 KMY92553.1 JH712088 EJD76154.1 AJVK01015207 AJVK01015208 AJVK01015209 GECU01036129 JAS71577.1 MWRG01001283 PRD34414.1 GBYB01005122 JAG74889.1 JYDL01000025 KRX23301.1 GAMC01016967 JAB89588.1 IACF01002329 LAB67987.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
UP000036403
+ More
UP000008237 UP000000311 UP000078541 UP000005203 UP000242457 UP000015104 UP000078542 UP000078200 UP000050793 UP000005205 UP000078540 UP000215335 UP000075809 UP000092444 UP000279307 UP000092460 UP000078492 UP000092445 UP000091820 UP000183832 UP000036681 UP000092461 UP000092443 UP000079169 UP000076858 UP000053825 UP000007875 UP000007266 UP000095301 UP000007819 UP000192223 UP000001555 UP000001292 UP000008144 UP000054359 UP000275408 UP000009046 UP000054843 UP000055024 UP000000803 UP000037069 UP000054632 UP000054805 UP000054826 UP000054815 UP000046394 UP000276776 UP000085678 UP000000304 UP000000305 UP000277928 UP000261560 UP000268350 UP000186922 UP000095285 UP000092462 UP000189705 UP000054630
UP000008237 UP000000311 UP000078541 UP000005203 UP000242457 UP000015104 UP000078542 UP000078200 UP000050793 UP000005205 UP000078540 UP000215335 UP000075809 UP000092444 UP000279307 UP000092460 UP000078492 UP000092445 UP000091820 UP000183832 UP000036681 UP000092461 UP000092443 UP000079169 UP000076858 UP000053825 UP000007875 UP000007266 UP000095301 UP000007819 UP000192223 UP000001555 UP000001292 UP000008144 UP000054359 UP000275408 UP000009046 UP000054843 UP000055024 UP000000803 UP000037069 UP000054632 UP000054805 UP000054826 UP000054815 UP000046394 UP000276776 UP000085678 UP000000304 UP000000305 UP000277928 UP000261560 UP000268350 UP000186922 UP000095285 UP000092462 UP000189705 UP000054630
PRIDE
Interpro
IPR002477
Peptidoglycan-bd-like
+ More
IPR024079 MetalloPept_cat_dom_sf
IPR036365 PGBD-like_sf
IPR028729 MMP15
IPR021158 Pept_M10A_Zn_BS
IPR001818 Pept_M10_metallopeptidase
IPR033739 M10A_MMP
IPR021190 Pept_M10A
IPR006026 Peptidase_Metallo
IPR036375 Hemopexin-like_dom_sf
IPR000585 Hemopexin-like_dom
IPR018487 Hemopexin-like_repeat
IPR028733 MMP25
IPR003582 ShKT_dom
IPR018486 Hemopexin_CS
IPR028726 MMP17
IPR028723 MMP24
IPR024079 MetalloPept_cat_dom_sf
IPR036365 PGBD-like_sf
IPR028729 MMP15
IPR021158 Pept_M10A_Zn_BS
IPR001818 Pept_M10_metallopeptidase
IPR033739 M10A_MMP
IPR021190 Pept_M10A
IPR006026 Peptidase_Metallo
IPR036375 Hemopexin-like_dom_sf
IPR000585 Hemopexin-like_dom
IPR018487 Hemopexin-like_repeat
IPR028733 MMP25
IPR003582 ShKT_dom
IPR018486 Hemopexin_CS
IPR028726 MMP17
IPR028723 MMP24
Gene 3D
ProteinModelPortal
A0A2A4IWM9
A0A2H1VB64
A0A194QU76
A0A194PQM8
A0A212FGS1
A0A0L7LT68
+ More
A0A336N0B2 A0A0J7L3C7 E9IWG0 A0A336MJP8 E2BGY0 E2AGJ9 A0A195FKY6 A0A087ZQ14 A0A1Y1LH94 A0A2A3EGV3 T1KNP2 A0A2S2NLZ5 A0A151IPY0 A0A0P5H5I2 A0A1A9V8N5 A0A183IHM5 A0A2H8TRK4 A0A158NET9 A0A195AV09 A0A348G5Y9 A0A232FL65 A0A151XIP8 A0A1B0G123 A0A3L8DAM3 A0A1B0ARG3 A0A195E8Z7 A0A1B0AD89 A0A1A9W7U8 A0A1J1I9A7 A0A0M3HUN9 A0A1B0GI40 A0A1A9YH87 A0A1S3DAR8 A0A0T6B4Y1 A0A164KHU5 A0A0L7R185 H2ZEY6 A0A1B6E667 D6WYY9 A0A147BPY1 A0A2S2Q4U3 T2M4Y7 A0A1I8MCL0 A0A0P4ZTH9 A0A0P5BVX1 A0A0P6BPJ7 X1WIM9 A0A1W4WJ61 B7QD72 B4HT26 A0A0P6IFT8 A0A0P6F6M4 F6RRU7 A0A0S3Q290 A0A1B6G040 A0A087USP2 A0A1W2W5Z1 A0A3M6V0B5 E0V8V4 A0A0V1N7S9 A0A0V1HWH9 Q8MPP3 E0VM71 A0A0L0BR13 A0A0V1EZ73 A0A0V1HWB5 A0A0V1JCY4 A0A0V0YC80 A0A0N5D346 A0A0V1EYX9 A0A0V1JD56 A0A1B0EX88 A0A1S3IGL5 T2M2R4 B4QHP4 Q9U9P0 E9GW38 H2ZEY7 A0A3P6UHJ2 A0A1W0XBN1 A0A3B3E0A3 A0A3B0JKN2 A0A0J9RA38 A0A1D1VJZ5 A0A0J9R8Z6 A0A1I7VM22 A0A1B0GPG4 A0A1B6HAB5 A0A2P6L761 A0A3Q0HIT9 A0A0C9QWS2 A0A0V0SA94 W8AXQ9 A0A2P2I1R8
A0A336N0B2 A0A0J7L3C7 E9IWG0 A0A336MJP8 E2BGY0 E2AGJ9 A0A195FKY6 A0A087ZQ14 A0A1Y1LH94 A0A2A3EGV3 T1KNP2 A0A2S2NLZ5 A0A151IPY0 A0A0P5H5I2 A0A1A9V8N5 A0A183IHM5 A0A2H8TRK4 A0A158NET9 A0A195AV09 A0A348G5Y9 A0A232FL65 A0A151XIP8 A0A1B0G123 A0A3L8DAM3 A0A1B0ARG3 A0A195E8Z7 A0A1B0AD89 A0A1A9W7U8 A0A1J1I9A7 A0A0M3HUN9 A0A1B0GI40 A0A1A9YH87 A0A1S3DAR8 A0A0T6B4Y1 A0A164KHU5 A0A0L7R185 H2ZEY6 A0A1B6E667 D6WYY9 A0A147BPY1 A0A2S2Q4U3 T2M4Y7 A0A1I8MCL0 A0A0P4ZTH9 A0A0P5BVX1 A0A0P6BPJ7 X1WIM9 A0A1W4WJ61 B7QD72 B4HT26 A0A0P6IFT8 A0A0P6F6M4 F6RRU7 A0A0S3Q290 A0A1B6G040 A0A087USP2 A0A1W2W5Z1 A0A3M6V0B5 E0V8V4 A0A0V1N7S9 A0A0V1HWH9 Q8MPP3 E0VM71 A0A0L0BR13 A0A0V1EZ73 A0A0V1HWB5 A0A0V1JCY4 A0A0V0YC80 A0A0N5D346 A0A0V1EYX9 A0A0V1JD56 A0A1B0EX88 A0A1S3IGL5 T2M2R4 B4QHP4 Q9U9P0 E9GW38 H2ZEY7 A0A3P6UHJ2 A0A1W0XBN1 A0A3B3E0A3 A0A3B0JKN2 A0A0J9RA38 A0A1D1VJZ5 A0A0J9R8Z6 A0A1I7VM22 A0A1B0GPG4 A0A1B6HAB5 A0A2P6L761 A0A3Q0HIT9 A0A0C9QWS2 A0A0V0SA94 W8AXQ9 A0A2P2I1R8
PDB
1SU3
E-value=9.43157e-05,
Score=102
Ontologies
GO
GO:0008237
GO:0031012
GO:0004222
GO:0008270
GO:0016021
GO:0030574
GO:0030198
GO:0005615
GO:0045216
GO:0016485
GO:0007426
GO:0040037
GO:0030728
GO:0009897
GO:0007561
GO:1901202
GO:0008045
GO:0097156
GO:0006929
GO:0034769
GO:0090090
GO:0007505
GO:0002218
GO:0030425
GO:0046529
GO:0003319
GO:0003144
GO:0035202
GO:0042060
GO:0007419
GO:0046331
GO:2000647
GO:0046872
GO:0005886
GO:0004175
GO:0071711
GO:0007424
GO:0006508
GO:0005089
GO:0035023
GO:0005515
GO:0016742
GO:0003824
Topology
Subcellular location
Cell membrane
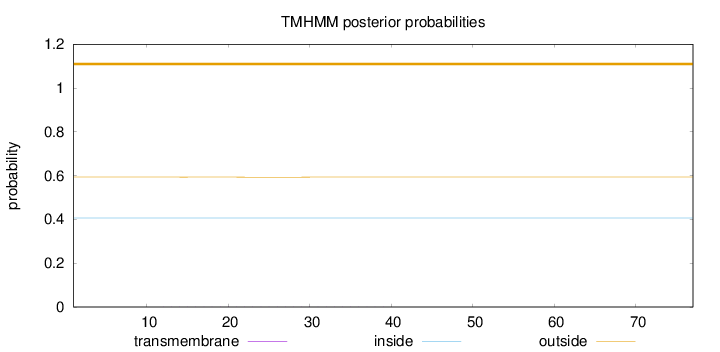
Length:
77
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00921
Exp number, first 60 AAs:
0.00921
Total prob of N-in:
0.40654
outside
1 - 77
Population Genetic Test Statistics
Pi
191.773493
Theta
169.378994
Tajima's D
0.417975
CLR
0.051958
CSRT
0.491825408729564
Interpretation
Uncertain
