Gene
KWMTBOMO04623
Pre Gene Modal
BGIBMGA005310
Annotation
PREDICTED:_matrix_metalloproteinase-14-like_[Plutella_xylostella]
Location in the cell
Extracellular Reliability : 1.46 Nuclear Reliability : 1.352
Sequence
CDS
ATGTTTTTGTCATCGACGCGTCGTCCTTCTCTACTGGACCCGTACGAGACACGCAGCGTGTTGGCACGCGCCCTCGACGTTTGGGAACAAGCCTCCCGACTTACTTTCACAGAAGTAAATTCGGAAGAGGCCGATATCCTTGTCTCGTTCGCCAAACGTTACCATGACGATGCATATCCATTCGATGGACGAGGTACGATTCTAGCTCACGCTTTCTTTCCTGGCGTGGACCGCGGTGGTGACGCGCACTTTGATGATGACGAACTCTGGTTGTTAGAGCCCAAAGATGACGATGAAGAAGGCACATCACTTTTTGCTGTTGCTGTTCACGAGTTTGGACATTCGTTAGGACTTAGCCACAGTAGTGTAAAAGGCGCACTGATGTACCCTTGGTATCAGGGTATTCAATCTAACTTTGTATTACCTGAAGATGATCGTAATGGCATTCAACAAATGTATGGTCCAAAATTAAACAATAAGAAATGGGCAAGAATCCCTTCATACAAACCAGTTCAAACACCACCAACAACCACTACGACTACCACTACGACCACAACAACCAGAAGGCCTTATATCCACCATAATAGACATCATGGAGTTCCGAACATGCCCCGTGATCCATCTAGAAACCCAGTTTACTACCCGAGAAGACCTTATTACCCTGATCGTCATTACCACAATACAACCGAAGATCATCCACGCAGAACGAATTACAATTACCCTCGAATTGAGACTACCACGCACGCAATAACTTACCGCCCCCGATACCCCTATAAACCGGAGTATCCCAGCTACCCAAGACCAAACTATCCGGTCGATCCTAGGCAAGATTACCCTAAGCAGGACTACCCTAGATCAGATTATCCTAAGCGACCATACTATCCAGAGAAAACTACCACTACTACTCAAAGGCCCACGCCGCCACAAAGGCCGACTCCAACTTCACCATCGGATAAACCAAATACGTGTGATACAAATTACGACGCTGTCGCTTTAATACGTGGCGAACTCTTCATATTCAAAGACAGGTATCACTGGAGGATCGGAGCTCACGGTCGATATCCTGGTTACCCAATGGAAATAACTAGAATGTGGTCCAGTTTACCAAAAAATCTAACACACGTGGACGCTGTTTATGAAAGACCAGATAAACATATTGCTATCTTTATAGGAAAGGAGTTGTATTTGTTCAATTCACAATATTTATTGCCCGGCTATCCGAAACCTTTGACCAAACTGGGTTTACCTGAAACTTTGGAGAAATTGGACGCGGCAATGGTTTGGAGTCTCAATGGCAAAACTTACTTTTACAGTGGAACTATGTATTGGAAATATGATGAAGACGAAGGAAAAGTAGAACTGGACTATCCCCGTAATATGGCCATGTGGAAGGGGGTTGGATACAACATCGATAGTGTTTTCCAATGGAAAGACGGGAACACCTATTTCTTCAAAGGAAAAGGATATTGGAAATTCAACGACCTGCAAATGCGAGTGGAGCACGAACAACAACTTCCGTCAGCCCCGTTCTGGATGGGTTGTCCTGGAGAGAGAAGCGGTCGAAAATATCCTTATCGCGCTCCTGCAGCACCAGGATCTACTTTACGATCGCCGAGTTCATCATCAACGCTACATTTAACTAACTTAATCGCACTTATCTCTTCTCTAGTTATTTCGCTCAATATGTCTTTACAATTTTACTGA
Protein
MFLSSTRRPSLLDPYETRSVLARALDVWEQASRLTFTEVNSEEADILVSFAKRYHDDAYPFDGRGTILAHAFFPGVDRGGDAHFDDDELWLLEPKDDDEEGTSLFAVAVHEFGHSLGLSHSSVKGALMYPWYQGIQSNFVLPEDDRNGIQQMYGPKLNNKKWARIPSYKPVQTPPTTTTTTTTTTTTRRPYIHHNRHHGVPNMPRDPSRNPVYYPRRPYYPDRHYHNTTEDHPRRTNYNYPRIETTTHAITYRPRYPYKPEYPSYPRPNYPVDPRQDYPKQDYPRSDYPKRPYYPEKTTTTTQRPTPPQRPTPTSPSDKPNTCDTNYDAVALIRGELFIFKDRYHWRIGAHGRYPGYPMEITRMWSSLPKNLTHVDAVYERPDKHIAIFIGKELYLFNSQYLLPGYPKPLTKLGLPETLEKLDAAMVWSLNGKTYFYSGTMYWKYDEDEGKVELDYPRNMAMWKGVGYNIDSVFQWKDGNTYFFKGKGYWKFNDLQMRVEHEQQLPSAPFWMGCPGERSGRKYPYRAPAAPGSTLRSPSSSSTLHLTNLIALISSLVISLNMSLQFY
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M10A family.
Uniprot
EMBL
Proteomes
PRIDE
Interpro
SUPFAM
SSF50923
SSF50923
Gene 3D
ProteinModelPortal
PDB
6CM1
E-value=8.53312e-39,
Score=404
Ontologies
GO
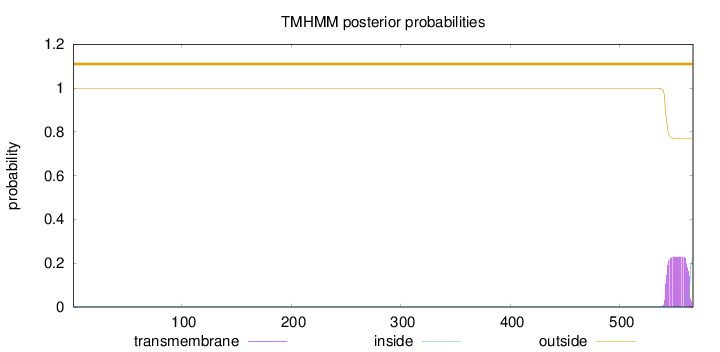
Topology
Length:
567
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.87145
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00223
outside
1 - 567
Population Genetic Test Statistics
Pi
163.796281
Theta
175.261382
Tajima's D
-0.364083
CLR
0.373427
CSRT
0.273286335683216
Interpretation
Uncertain
