Pre Gene Modal
BGIBMGA005499
Annotation
PREDICTED:_transmembrane_9_superfamily_member_3_[Papilio_xuthus]
Full name
Transmembrane 9 superfamily member
Location in the cell
PlasmaMembrane Reliability : 4.94
Sequence
CDS
ATGAAATTTTTATATATATTGTTTATAATGTGGACTATAGTTCATTGCGATGAACACACACATTCCTATAAAGATGGAGAACAAGTAGTGCTTTGGATGAACACCGTGGGTCCGTACCATAACCGACAAGAGACATATGCCTATTTTTCGCTCCCGTTTTGCGTTGGCACTAAAGTCACTATTGGACATTATCATGAAACTCTATCTGAAGCTCTACAAGGCGTCGAATTGGAGTTCAGCGGTCTTGATATTACGTTTAAGGAAAATGTGCCGGCGCAGCAATTTTGTGCCATCGAACTCAACGATCAAACTTACAAAGCTTTCGTTTATGCTGTCAAAAACCATTATTGGTATCAAATGTATATAGACGATCTGCCAATATGGGGCACTGTAGGTGAGATTGATGGAGACCATTTTTACATCTGGACTCACAAGAAATTTGACATTGGGTATAATGGAAACAGGATTGTTGAAGTCAACCTTACAGCAGAAAACAAAGAACGGCTTAGTCCTGATGCAAAGATCCCATTTACATATGAAGTTAACTGGAAGAAGAGCAACATAAATTTTGAAGATCGATTTGATAAGTACTTAGATCCAAACTTTTTTCAACACCGAATTCATTGGTTCAGTATTTTCAATAGCTTCATGATGGTTATATTTTTAGTTGGTCTTGTTTCTATGATACTCATGAGAACTCTTCGAAAAGATTATGCTAGATATTCCAAAGATGATGATCTTGATGATTTGGAAAAGGATCTAGGTGATGAATATGGTTGGAAACAGGTTCATGGGGATGTATTCAGACCTGTACCACATTTATCTTTATTTTCAGCATTAGTTGGTTCTGGCTATCAATTGACTGTAGTAACTTTGGCAGTTATTATTTTCACTATTTTTGGAGAATTGTATACTGAAAGGGGCTCACTCTTATCAACTGCAATCTTTATTTATGCAGCTACATCACCAGTCAATGGTTATTTTGGAGGATCCCTGTATTCTCGAATGGGTGGTAAATTGTGGATAAAACAAATGTTGTTGTCAGCATTTTTCTTACCAGTCTTAGTTTGTGGGACTGCCTTTTTTATTAACTTTATTGCTATGTACTATCATGCTTCTAGAGCTATTCCTTTTGGCAGTATGATTGCTGTGATGTCTATCTGCACTTTTGTGATTTTACCATTAACTCTTGTTGGAACGGTACTTGGAAGGAATTTAGCTGGACAACCAGATTACCCTTGTAGAATCAATGCAGTTCCAAGGCCTATTCCAGAGAAGAAGTGGTTTATGGAACCTCTAATAATCATTATTGTTGGAGGAGTATTACCATTTGGATCTATCTTTATTGAAATGTACTTTATCTTTACATCATTCTGGGCATACAAGATCTACTATGTGTATGGGTTTATGTTGCTAGTGTTCTTAATATTGATGATTGTAACTGTATGTGTCACTATTGTTTGTACTTATTTCTTGTTGAATGCTGAAGATTACCGCTGGCAGTGGACCAGTTTCTTATCTGCAGGGAGCACTGCAGTTTATGTGTATTTATATTCATTTTACTATTTCTTGTTTAAAACAAAGATGTATGGTTTATTTCAAACAACATTTTATTTTGGCTATATGGCTTTGTTCAGTTTAGCTTTAGGCATTATTTGTGGGACAGTTGGCTATGTAGGCACAAGCATATTTGTACGTAAAATTTACTCTACAGTAAAAATTGATTAA
Protein
MKFLYILFIMWTIVHCDEHTHSYKDGEQVVLWMNTVGPYHNRQETYAYFSLPFCVGTKVTIGHYHETLSEALQGVELEFSGLDITFKENVPAQQFCAIELNDQTYKAFVYAVKNHYWYQMYIDDLPIWGTVGEIDGDHFYIWTHKKFDIGYNGNRIVEVNLTAENKERLSPDAKIPFTYEVNWKKSNINFEDRFDKYLDPNFFQHRIHWFSIFNSFMMVIFLVGLVSMILMRTLRKDYARYSKDDDLDDLEKDLGDEYGWKQVHGDVFRPVPHLSLFSALVGSGYQLTVVTLAVIIFTIFGELYTERGSLLSTAIFIYAATSPVNGYFGGSLYSRMGGKLWIKQMLLSAFFLPVLVCGTAFFINFIAMYYHASRAIPFGSMIAVMSICTFVILPLTLVGTVLGRNLAGQPDYPCRINAVPRPIPEKKWFMEPLIIIIVGGVLPFGSIFIEMYFIFTSFWAYKIYYVYGFMLLVFLILMIVTVCVTIVCTYFLLNAEDYRWQWTSFLSAGSTAVYVYLYSFYYFLFKTKMYGLFQTTFYFGYMALFSLALGIICGTVGYVGTSIFVRKIYSTVKID
Summary
Similarity
Belongs to the nonaspanin (TM9SF) (TC 9.A.2) family.
Feature
chain Transmembrane 9 superfamily member
Uniprot
H9J7K7
A0A1E1W8F6
A0A0L7KYT7
A0A2H1V0H2
A0A194PRN0
A0A194QST2
+ More
A0A2J7RQE2 A0A2P8YC66 A0A1S3HHU1 A0A1B6ET88 A0A1B6E679 E2B9M3 A0A026WXV1 A0A1B6J1P4 A0A195B981 A0A3L8DBP6 F4WNZ1 A0A158NPG8 A0A195E5G3 A0A0B7AVE5 A0A195ETX5 E2AJW1 V5GID1 V9ILI2 A0A1Y1KHT4 A0A154PFZ6 J3JYQ3 A0A0C9RHK9 K7J157 N6TXS1 A0A023EWL1 A0A1Q3G2X5 A0A2I9LPU1 A0A2C9JFH9 A0A1W7R9E4 D6WFQ6 Q16NC8 Q16KY5 A0A088AA94 A0A0M8ZU53 A0A232EX25 A0A195CMB8 A0A0L7QQG5 A0A310SHU1 U5EV54 R7U229 A0A336LDW9 A0A182Q6Z5 V3ZQV1 A0A182NNJ2 A0A2M4DMN2 A0A087U7P9 A0A182LI01 A0A182RKJ2 A0A2M4A259 A0A182UZC3 A0A2M4A216 A0A182FDZ9 A0A2M3Z086 K1PSA1 A0A1W4WKF5 A0A182JP98 A0A0V0G3Q5 A0A069DVV3 A0A0N7Z9E8 T1HIT0 A0A2R5LIF5 C3Z4N8 A0A224XM66 A0A151WRB5 A0A2L2Y4W7 A0A293MVL2 A0A131Y1V3 A0A0K8R436 B7QBL1 T1DI10 A0A1L8DUP0 A0A0L8FKQ8 A0A1I8MRV4 A0A1B0G264 A0A1E1X9I9 A0A067R0M4 A0A224ZAL1 A0A131Z0U6 L7M080 A0A1I8Q961 A0A1A9XJI5 A0A1A9UJ79 A0A1B0A6L0 A0A3S0ZMV3 A0A2A3E158 A0A023FNJ3 A0A2T7PWR3 A0A1A9X5Q9 A0A0L0CPN2 A0A3P8VQZ4 A0A060W1U6 A8E7G8 Q7ZV33 A0A2D4JP52
A0A2J7RQE2 A0A2P8YC66 A0A1S3HHU1 A0A1B6ET88 A0A1B6E679 E2B9M3 A0A026WXV1 A0A1B6J1P4 A0A195B981 A0A3L8DBP6 F4WNZ1 A0A158NPG8 A0A195E5G3 A0A0B7AVE5 A0A195ETX5 E2AJW1 V5GID1 V9ILI2 A0A1Y1KHT4 A0A154PFZ6 J3JYQ3 A0A0C9RHK9 K7J157 N6TXS1 A0A023EWL1 A0A1Q3G2X5 A0A2I9LPU1 A0A2C9JFH9 A0A1W7R9E4 D6WFQ6 Q16NC8 Q16KY5 A0A088AA94 A0A0M8ZU53 A0A232EX25 A0A195CMB8 A0A0L7QQG5 A0A310SHU1 U5EV54 R7U229 A0A336LDW9 A0A182Q6Z5 V3ZQV1 A0A182NNJ2 A0A2M4DMN2 A0A087U7P9 A0A182LI01 A0A182RKJ2 A0A2M4A259 A0A182UZC3 A0A2M4A216 A0A182FDZ9 A0A2M3Z086 K1PSA1 A0A1W4WKF5 A0A182JP98 A0A0V0G3Q5 A0A069DVV3 A0A0N7Z9E8 T1HIT0 A0A2R5LIF5 C3Z4N8 A0A224XM66 A0A151WRB5 A0A2L2Y4W7 A0A293MVL2 A0A131Y1V3 A0A0K8R436 B7QBL1 T1DI10 A0A1L8DUP0 A0A0L8FKQ8 A0A1I8MRV4 A0A1B0G264 A0A1E1X9I9 A0A067R0M4 A0A224ZAL1 A0A131Z0U6 L7M080 A0A1I8Q961 A0A1A9XJI5 A0A1A9UJ79 A0A1B0A6L0 A0A3S0ZMV3 A0A2A3E158 A0A023FNJ3 A0A2T7PWR3 A0A1A9X5Q9 A0A0L0CPN2 A0A3P8VQZ4 A0A060W1U6 A8E7G8 Q7ZV33 A0A2D4JP52
Pubmed
19121390
26227816
26354079
29403074
20798317
24508170
+ More
30249741 21719571 21347285 28004739 22516182 20075255 23537049 24945155 29248469 15562597 18362917 19820115 17510324 28648823 23254933 20966253 22992520 26334808 27129103 18563158 26561354 24330624 25315136 28503490 24845553 28797301 26830274 25576852 26108605 24487278 24755649 23594743
30249741 21719571 21347285 28004739 22516182 20075255 23537049 24945155 29248469 15562597 18362917 19820115 17510324 28648823 23254933 20966253 22992520 26334808 27129103 18563158 26561354 24330624 25315136 28503490 24845553 28797301 26830274 25576852 26108605 24487278 24755649 23594743
EMBL
BABH01013963
GDQN01007772
JAT83282.1
JTDY01004221
KOB68438.1
ODYU01000113
+ More
SOQ34337.1 KQ459595 KPI95608.1 KQ461154 KPJ08537.1 NEVH01001340 PNF43049.1 PYGN01000711 PSN41847.1 GECZ01028632 JAS41137.1 GEDC01029412 GEDC01014129 GEDC01008228 GEDC01003901 JAS07886.1 JAS23169.1 JAS29070.1 JAS33397.1 GL446573 EFN87604.1 KK107077 QOIP01000010 EZA60571.1 RLU18092.1 GECU01014666 JAS93040.1 KQ976542 KYM81098.1 RLU17751.1 GL888243 EGI63922.1 ADTU01022529 KQ979608 KYN20326.1 HACG01037973 CEK84838.1 KQ981965 KYN31698.1 GL440100 EFN66277.1 GALX01004647 JAB63819.1 JR049989 AEY61149.1 GEZM01086079 GEZM01086078 JAV59790.1 KQ434889 KZC10238.1 BT128383 AEE63341.1 GBYB01006416 JAG76183.1 APGK01050932 KB741178 KB632325 ENN73181.1 ERL92379.1 GAPW01000679 GEHC01000356 JAC12919.1 JAV47289.1 GFDL01000878 JAV34167.1 GFWZ01000416 MBW20406.1 GFAH01000627 JAV47762.1 KQ971319 EEZ99577.2 CH477829 EAT35846.1 CH477932 EAT34964.1 KQ435878 KOX70046.1 NNAY01001794 OXU22901.1 KQ977574 KYN01835.1 KQ414786 KOC60858.1 KQ765605 OAD53865.1 GANO01002015 JAB57856.1 AMQN01009728 KB306072 ELU00390.1 UFQS01003941 UFQS01003963 UFQT01003941 UFQT01003963 SSX16046.1 SSX16063.1 SSX35379.1 AXCN02001915 KB203382 ESO84885.1 GGFL01014636 MBW78814.1 KK118595 KFM73388.1 GGFK01001518 MBW34839.1 GGFK01001478 MBW34799.1 GGFM01001155 MBW21906.1 JH816628 EKC24508.1 GECL01003441 JAP02683.1 GBGD01000938 JAC87951.1 GDKW01000747 JAI55848.1 ACPB03009657 GGLE01004981 MBY09107.1 GG666580 EEN52477.1 GFTR01007327 JAW09099.1 KQ982807 KYQ50406.1 IAAA01009326 IAAA01009327 LAA02340.1 GFWV01020146 MAA44874.1 GEFM01003925 JAP71871.1 GADI01008190 JAA65618.1 ABJB010062463 ABJB010412400 ABJB010611035 ABJB010702141 DS902191 EEC16233.1 GALA01001211 JAA93641.1 GFDF01003933 JAV10151.1 KQ429656 KOF65281.1 CCAG010019770 GFAC01003286 JAT95902.1 KK853125 KDR11018.1 GFPF01012768 MAA23914.1 GEDV01004202 JAP84355.1 GACK01007308 JAA57726.1 RQTK01000274 RUS82727.1 KZ288466 PBC25445.1 GBBK01001658 JAC22824.1 PZQS01000001 PVD37830.1 JRES01000089 KNC34221.1 FR904367 CDQ61092.1 BX004975 BX248127 BC046021 AAH46021.1 IACK01231287 LAA98174.1
SOQ34337.1 KQ459595 KPI95608.1 KQ461154 KPJ08537.1 NEVH01001340 PNF43049.1 PYGN01000711 PSN41847.1 GECZ01028632 JAS41137.1 GEDC01029412 GEDC01014129 GEDC01008228 GEDC01003901 JAS07886.1 JAS23169.1 JAS29070.1 JAS33397.1 GL446573 EFN87604.1 KK107077 QOIP01000010 EZA60571.1 RLU18092.1 GECU01014666 JAS93040.1 KQ976542 KYM81098.1 RLU17751.1 GL888243 EGI63922.1 ADTU01022529 KQ979608 KYN20326.1 HACG01037973 CEK84838.1 KQ981965 KYN31698.1 GL440100 EFN66277.1 GALX01004647 JAB63819.1 JR049989 AEY61149.1 GEZM01086079 GEZM01086078 JAV59790.1 KQ434889 KZC10238.1 BT128383 AEE63341.1 GBYB01006416 JAG76183.1 APGK01050932 KB741178 KB632325 ENN73181.1 ERL92379.1 GAPW01000679 GEHC01000356 JAC12919.1 JAV47289.1 GFDL01000878 JAV34167.1 GFWZ01000416 MBW20406.1 GFAH01000627 JAV47762.1 KQ971319 EEZ99577.2 CH477829 EAT35846.1 CH477932 EAT34964.1 KQ435878 KOX70046.1 NNAY01001794 OXU22901.1 KQ977574 KYN01835.1 KQ414786 KOC60858.1 KQ765605 OAD53865.1 GANO01002015 JAB57856.1 AMQN01009728 KB306072 ELU00390.1 UFQS01003941 UFQS01003963 UFQT01003941 UFQT01003963 SSX16046.1 SSX16063.1 SSX35379.1 AXCN02001915 KB203382 ESO84885.1 GGFL01014636 MBW78814.1 KK118595 KFM73388.1 GGFK01001518 MBW34839.1 GGFK01001478 MBW34799.1 GGFM01001155 MBW21906.1 JH816628 EKC24508.1 GECL01003441 JAP02683.1 GBGD01000938 JAC87951.1 GDKW01000747 JAI55848.1 ACPB03009657 GGLE01004981 MBY09107.1 GG666580 EEN52477.1 GFTR01007327 JAW09099.1 KQ982807 KYQ50406.1 IAAA01009326 IAAA01009327 LAA02340.1 GFWV01020146 MAA44874.1 GEFM01003925 JAP71871.1 GADI01008190 JAA65618.1 ABJB010062463 ABJB010412400 ABJB010611035 ABJB010702141 DS902191 EEC16233.1 GALA01001211 JAA93641.1 GFDF01003933 JAV10151.1 KQ429656 KOF65281.1 CCAG010019770 GFAC01003286 JAT95902.1 KK853125 KDR11018.1 GFPF01012768 MAA23914.1 GEDV01004202 JAP84355.1 GACK01007308 JAA57726.1 RQTK01000274 RUS82727.1 KZ288466 PBC25445.1 GBBK01001658 JAC22824.1 PZQS01000001 PVD37830.1 JRES01000089 KNC34221.1 FR904367 CDQ61092.1 BX004975 BX248127 BC046021 AAH46021.1 IACK01231287 LAA98174.1
Proteomes
UP000005204
UP000037510
UP000053268
UP000053240
UP000235965
UP000245037
+ More
UP000085678 UP000008237 UP000053097 UP000279307 UP000078540 UP000007755 UP000005205 UP000078492 UP000078541 UP000000311 UP000076502 UP000002358 UP000019118 UP000030742 UP000076420 UP000007266 UP000008820 UP000005203 UP000053105 UP000215335 UP000078542 UP000053825 UP000014760 UP000075886 UP000030746 UP000075884 UP000054359 UP000075882 UP000075900 UP000075903 UP000069272 UP000005408 UP000192223 UP000075881 UP000015103 UP000001554 UP000075809 UP000001555 UP000053454 UP000095301 UP000092444 UP000027135 UP000095300 UP000092443 UP000078200 UP000092445 UP000271974 UP000242457 UP000245119 UP000091820 UP000037069 UP000265120 UP000193380 UP000000437
UP000085678 UP000008237 UP000053097 UP000279307 UP000078540 UP000007755 UP000005205 UP000078492 UP000078541 UP000000311 UP000076502 UP000002358 UP000019118 UP000030742 UP000076420 UP000007266 UP000008820 UP000005203 UP000053105 UP000215335 UP000078542 UP000053825 UP000014760 UP000075886 UP000030746 UP000075884 UP000054359 UP000075882 UP000075900 UP000075903 UP000069272 UP000005408 UP000192223 UP000075881 UP000015103 UP000001554 UP000075809 UP000001555 UP000053454 UP000095301 UP000092444 UP000027135 UP000095300 UP000092443 UP000078200 UP000092445 UP000271974 UP000242457 UP000245119 UP000091820 UP000037069 UP000265120 UP000193380 UP000000437
PRIDE
Interpro
SUPFAM
SSF52799
SSF52799
Gene 3D
ProteinModelPortal
H9J7K7
A0A1E1W8F6
A0A0L7KYT7
A0A2H1V0H2
A0A194PRN0
A0A194QST2
+ More
A0A2J7RQE2 A0A2P8YC66 A0A1S3HHU1 A0A1B6ET88 A0A1B6E679 E2B9M3 A0A026WXV1 A0A1B6J1P4 A0A195B981 A0A3L8DBP6 F4WNZ1 A0A158NPG8 A0A195E5G3 A0A0B7AVE5 A0A195ETX5 E2AJW1 V5GID1 V9ILI2 A0A1Y1KHT4 A0A154PFZ6 J3JYQ3 A0A0C9RHK9 K7J157 N6TXS1 A0A023EWL1 A0A1Q3G2X5 A0A2I9LPU1 A0A2C9JFH9 A0A1W7R9E4 D6WFQ6 Q16NC8 Q16KY5 A0A088AA94 A0A0M8ZU53 A0A232EX25 A0A195CMB8 A0A0L7QQG5 A0A310SHU1 U5EV54 R7U229 A0A336LDW9 A0A182Q6Z5 V3ZQV1 A0A182NNJ2 A0A2M4DMN2 A0A087U7P9 A0A182LI01 A0A182RKJ2 A0A2M4A259 A0A182UZC3 A0A2M4A216 A0A182FDZ9 A0A2M3Z086 K1PSA1 A0A1W4WKF5 A0A182JP98 A0A0V0G3Q5 A0A069DVV3 A0A0N7Z9E8 T1HIT0 A0A2R5LIF5 C3Z4N8 A0A224XM66 A0A151WRB5 A0A2L2Y4W7 A0A293MVL2 A0A131Y1V3 A0A0K8R436 B7QBL1 T1DI10 A0A1L8DUP0 A0A0L8FKQ8 A0A1I8MRV4 A0A1B0G264 A0A1E1X9I9 A0A067R0M4 A0A224ZAL1 A0A131Z0U6 L7M080 A0A1I8Q961 A0A1A9XJI5 A0A1A9UJ79 A0A1B0A6L0 A0A3S0ZMV3 A0A2A3E158 A0A023FNJ3 A0A2T7PWR3 A0A1A9X5Q9 A0A0L0CPN2 A0A3P8VQZ4 A0A060W1U6 A8E7G8 Q7ZV33 A0A2D4JP52
A0A2J7RQE2 A0A2P8YC66 A0A1S3HHU1 A0A1B6ET88 A0A1B6E679 E2B9M3 A0A026WXV1 A0A1B6J1P4 A0A195B981 A0A3L8DBP6 F4WNZ1 A0A158NPG8 A0A195E5G3 A0A0B7AVE5 A0A195ETX5 E2AJW1 V5GID1 V9ILI2 A0A1Y1KHT4 A0A154PFZ6 J3JYQ3 A0A0C9RHK9 K7J157 N6TXS1 A0A023EWL1 A0A1Q3G2X5 A0A2I9LPU1 A0A2C9JFH9 A0A1W7R9E4 D6WFQ6 Q16NC8 Q16KY5 A0A088AA94 A0A0M8ZU53 A0A232EX25 A0A195CMB8 A0A0L7QQG5 A0A310SHU1 U5EV54 R7U229 A0A336LDW9 A0A182Q6Z5 V3ZQV1 A0A182NNJ2 A0A2M4DMN2 A0A087U7P9 A0A182LI01 A0A182RKJ2 A0A2M4A259 A0A182UZC3 A0A2M4A216 A0A182FDZ9 A0A2M3Z086 K1PSA1 A0A1W4WKF5 A0A182JP98 A0A0V0G3Q5 A0A069DVV3 A0A0N7Z9E8 T1HIT0 A0A2R5LIF5 C3Z4N8 A0A224XM66 A0A151WRB5 A0A2L2Y4W7 A0A293MVL2 A0A131Y1V3 A0A0K8R436 B7QBL1 T1DI10 A0A1L8DUP0 A0A0L8FKQ8 A0A1I8MRV4 A0A1B0G264 A0A1E1X9I9 A0A067R0M4 A0A224ZAL1 A0A131Z0U6 L7M080 A0A1I8Q961 A0A1A9XJI5 A0A1A9UJ79 A0A1B0A6L0 A0A3S0ZMV3 A0A2A3E158 A0A023FNJ3 A0A2T7PWR3 A0A1A9X5Q9 A0A0L0CPN2 A0A3P8VQZ4 A0A060W1U6 A8E7G8 Q7ZV33 A0A2D4JP52
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.998678
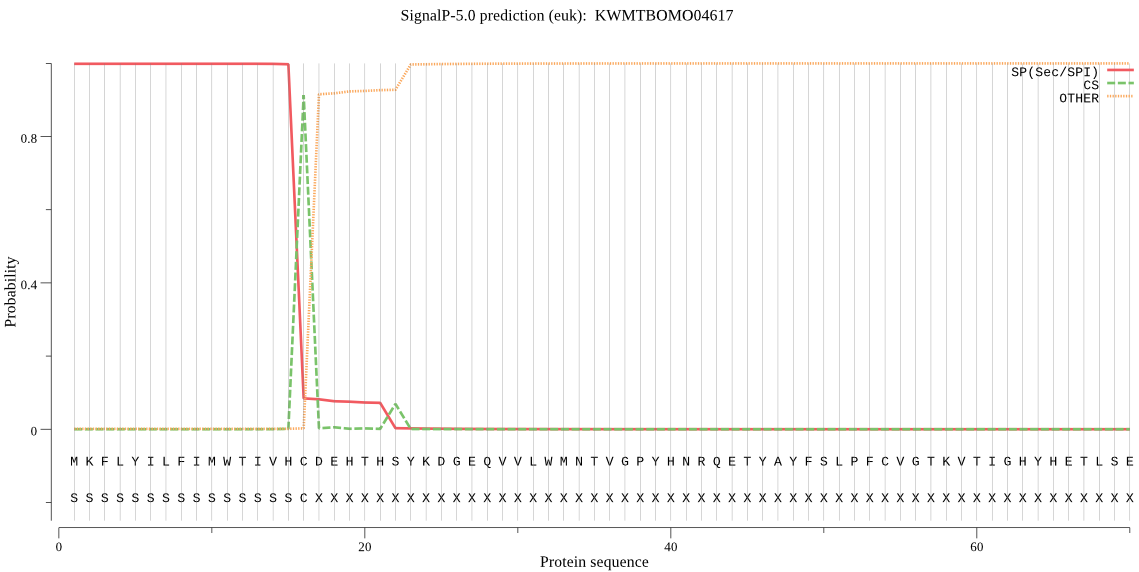
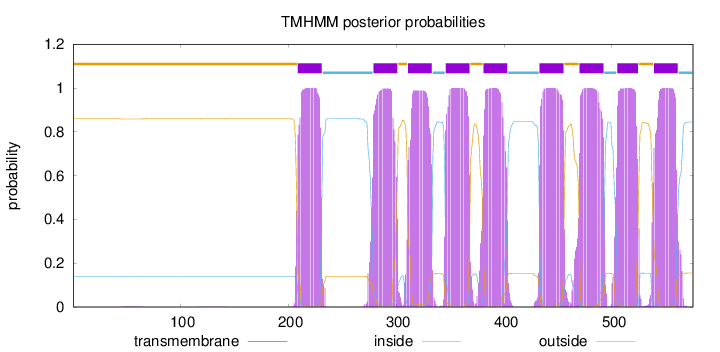
Length:
575
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
204.92801
Exp number, first 60 AAs:
0.02143
Total prob of N-in:
0.14069
outside
1 - 208
TMhelix
209 - 231
inside
232 - 278
TMhelix
279 - 301
outside
302 - 310
TMhelix
311 - 333
inside
334 - 345
TMhelix
346 - 368
outside
369 - 380
TMhelix
381 - 403
inside
404 - 432
TMhelix
433 - 455
outside
456 - 469
TMhelix
470 - 492
inside
493 - 504
TMhelix
505 - 524
outside
525 - 538
TMhelix
539 - 561
inside
562 - 575
Population Genetic Test Statistics
Pi
165.289767
Theta
189.822713
Tajima's D
-0.356105
CLR
183.287855
CSRT
0.276886155692215
Interpretation
Uncertain
