Pre Gene Modal
BGIBMGA005312
Annotation
PREDICTED:_notchless_protein_homolog_1_[Amyelois_transitella]
Full name
Notchless protein homolog 1
+ More
Protein Notchless
Protein Notchless
Location in the cell
Cytoplasmic Reliability : 1.394 Extracellular Reliability : 1.669
Sequence
CDS
ATGGAGGTAGATTCACCGAGCTGTATTCAAGCTAAATTTAAATCTGACACTGGAGAAGAAACGGGAAGTCCATTGGATTTGCCCCTAAATATAACCAAAGATCAACTTCAACTGATATGTAATGCACTTTTACAAGAGGAAGATAAACCATTCCTCTTTTTTGTGAAAGATGTTGAAATAACAACCACAATAAAGGATGCATTGGACCTAAAGACCTTGAATAGTGAAGAGGTTGTAGAGATAATATATCAACAACAGGCTGTATTTAGAGTTAGACCAGTCACAAGGTGTACTAGTTCAATACCAGGTCATGCGGAAGCAGTGATATCTACGTGCTTTAGCCCTTCCGGGAAGAAACTGGCTAGTGGCTCAGGTGACACTACTGTCAGATTTTGGGATCTGAACACACAGACACCTCTACATGTATGTAAAGGACATAGTAATTGGGTACTCTGTATAAGTTGGTCGCCAGATGGAAAAAAACTGGCATCAGCATGCAAACAAGGAAGAATTATTTTATGGGATCCATCAACTGGTAATCAAATTGGTAAACCTATGCTTGGACACAAACAGTGGGTAACAGCTCTGGCCTGGGAACCTTACCACAGAAACCCAGAATGTAGAAAACTTGCCAGTTCATCCAAAGATGGTGATGTTAGGATATGGGACACGGTAACATGTCTAACATTGTTGAGCTTGACTGGACATTCCAAAGCTGTAACTTCGGTGAAATGGGGTGGTAATGGTTTAATCTACACATCTTCTCAGGACAGGACAATCAAGGTGTGGAGGGCAGATGATGGGGTGTTATGCAGGACATTAGAAGGTCATGCTCATTGGGTTAACACTTTAGCACTAAACACGGATTATGTGTTACGTACTGGACCTTTTCATCCTGTCATGGACAATAACGCATATAGTGATGATAAGAATGTTCTCCAGCAAAGAGCATTGGACAGGTATAATGCAGTGTGCCAACAAGGAGCTGAACGTTTAGTTTCTGGTTCAGATGATTTTACATTATTTCTCTGGCTACCAGAAAAAGAAAAGAAGCCTCTGGCTAGAATGACTGGACATCAACAACTTATAAATGATGTGAAATTTTCACCAGATACAAGGATTATAGCGTCTGCTTCATTTGACAAATCTGTGAAATTATGGGAAGCAGCAAGTGGGAAGTTTATAACAACATTAAGGGGACATGTTCAAGCTGTATACATGGTTGCATGGTCTGCAGATTCTAGACTACTATTAAGTTCTAGTGCTGATTCTACACTGAAAGTTTGGAGTATGAAAACAAAGAAGTTAGAGTTGGATCTTCCAGGTCATGCAGATGAAGTGTATGCTGTCGATTGGTCCCCAGATGGAGCTTATGTGGCATCAGGAGGAAAAGATAAAGTGTTGAAGTTGTGGCAGCATTGA
Protein
MEVDSPSCIQAKFKSDTGEETGSPLDLPLNITKDQLQLICNALLQEEDKPFLFFVKDVEITTTIKDALDLKTLNSEEVVEIIYQQQAVFRVRPVTRCTSSIPGHAEAVISTCFSPSGKKLASGSGDTTVRFWDLNTQTPLHVCKGHSNWVLCISWSPDGKKLASACKQGRIILWDPSTGNQIGKPMLGHKQWVTALAWEPYHRNPECRKLASSSKDGDVRIWDTVTCLTLLSLTGHSKAVTSVKWGGNGLIYTSSQDRTIKVWRADDGVLCRTLEGHAHWVNTLALNTDYVLRTGPFHPVMDNNAYSDDKNVLQQRALDRYNAVCQQGAERLVSGSDDFTLFLWLPEKEKKPLARMTGHQQLINDVKFSPDTRIIASASFDKSVKLWEAASGKFITTLRGHVQAVYMVAWSADSRLLLSSSADSTLKVWSMKTKKLELDLPGHADEVYAVDWSPDGAYVASGGKDKVLKLWQH
Summary
Description
Plays a role in regulating the expression of CDKN1A and several members of the Wnt pathway, probably via its effects on Notch activity. Required for normal embryogenesis (By similarity). Plays a role in regulating Notch activity.
Plays a role in regulating Notch activity. Plays a role in regulating the expression of CDKN1A and several members of the Wnt pathway, probably via its effects on Notch activity. Required during embryogenesis for inner mass cell survival (By similarity).
Plays a role in regulating Notch activity.
Plays a role in regulating Notch activity. Plays a role in regulating the expression of CDKN1A and several members of the Wnt pathway, probably via its effects on Notch activity. Required during embryogenesis for inner mass cell survival (By similarity).
Plays a role in regulating Notch activity.
Subunit
Associates with the pre-60S ribosomal particle. Interacts (via WD repeats) with uL18 (By similarity). Interacts (via UBL domain) with MDN1 (via VWFA/MIDAS domain) (By similarity).
Interacts with Notch (via cytoplasmic domain) (PubMed:9857191). Associates with the pre-60S ribosomal particle (By similarity).
Associates with the pre-60S ribosomal particle. Interacts (via WD repeats) with uL18 (By similarity). Interacts (via UBL domain) with MDN1 (via VWFA/MIDAS domain) (PubMed:26601951).
Interacts with Notch (via cytoplasmic domain) (PubMed:9857191). Associates with the pre-60S ribosomal particle (By similarity).
Associates with the pre-60S ribosomal particle. Interacts (via WD repeats) with uL18 (By similarity). Interacts (via UBL domain) with MDN1 (via VWFA/MIDAS domain) (PubMed:26601951).
Miscellaneous
The name 'Notchless' derives from the ability to suppress the wing notching caused by some Notch mutant alleles.
Similarity
Belongs to the NLE1/RSA4 family.
Keywords
Notch signaling pathway
Nucleus
Repeat
WD repeat
Complete proteome
Phosphoprotein
Reference proteome
Acetylation
Alternative splicing
Direct protein sequencing
Polymorphism
Feature
chain Notchless protein homolog 1
splice variant In isoform 2.
sequence variant In dbSNP:rs1471615.
splice variant In isoform 2.
sequence variant In dbSNP:rs1471615.
Uniprot
H9J720
A0A2H1V0Q1
A0A1E1WD31
A0A3S2TNH8
A0A2A4J2B7
A0A194PQM6
+ More
A0A194QY83 S4NNP5 A0A212EYS4 A0A0T6B410 A0A1Y1JU29 A0A1W4WF11 A0A067QEW1 A0A2J7QXN3 A0A1B6CNS1 U4U3B9 A0A0P5B0P2 A0A0P6CUL8 A0A0P5E6A5 A0A0K8U036 A0A0N7ZQD1 A0A0P5ZCM1 E9HFT7 A0A0P6CI94 A0A034WDM4 A0A0K8WFQ0 A0A1B6GE01 W8BDS3 D6X557 A0A154PV51 K7J2R3 A0A1I8NEG9 A0A1S3D021 T1PIJ4 A0A0U2K782 A0A0V0G8D6 A0A146LXZ6 A0A182FB76 A0A1L8H809 A0A0N0BH53 A0A232F1K0 Q7ZXK9 A0A1A9VBQ2 A0A2M4BLN3 A0A1I8NN00 A0A0L0BXN3 A0A1B0G8B6 A0A023F589 E0VIV5 Q0D2B0 A0A151IFH4 W5JU99 A0A0C9S352 A0A2M4BLM9 B4KFG7 F4WZK7 A0A1A9WN04 Q58D20 A0A3Q1N213 I3M7E4 A0A0D9R1W1 A0A0J9TCL7 A0A195DXP8 A0A1A9XXQ7 B4P2G2 B3N7S2 Q9VPR4 H0X1H4 B4Q658 J9JTX5 A0A195FEH4 A0A091DT08 A0A1S3JDQ8 B3MLP8 A0A2K6QQG7 A0A2I2V019 A0A1S3W3K9 F7GHQ8 G3VN49 A0A3Q0DL33 U3DX50 E9ILD0 Q9NVX2 H0VN00 A0A2K6LFA1 A0A287A8X1 M3VUD4 A0A2K5RDR6 G2HGH5 A0A1D5R0W2 B4ICZ0 V9KP55 A0A2K5MYU0 A0A1B0APF9 A0A287ACH7 A0A096P3W2 A0A182VBH9 A0A182HZJ3 Q7PTK2 B2GV82
A0A194QY83 S4NNP5 A0A212EYS4 A0A0T6B410 A0A1Y1JU29 A0A1W4WF11 A0A067QEW1 A0A2J7QXN3 A0A1B6CNS1 U4U3B9 A0A0P5B0P2 A0A0P6CUL8 A0A0P5E6A5 A0A0K8U036 A0A0N7ZQD1 A0A0P5ZCM1 E9HFT7 A0A0P6CI94 A0A034WDM4 A0A0K8WFQ0 A0A1B6GE01 W8BDS3 D6X557 A0A154PV51 K7J2R3 A0A1I8NEG9 A0A1S3D021 T1PIJ4 A0A0U2K782 A0A0V0G8D6 A0A146LXZ6 A0A182FB76 A0A1L8H809 A0A0N0BH53 A0A232F1K0 Q7ZXK9 A0A1A9VBQ2 A0A2M4BLN3 A0A1I8NN00 A0A0L0BXN3 A0A1B0G8B6 A0A023F589 E0VIV5 Q0D2B0 A0A151IFH4 W5JU99 A0A0C9S352 A0A2M4BLM9 B4KFG7 F4WZK7 A0A1A9WN04 Q58D20 A0A3Q1N213 I3M7E4 A0A0D9R1W1 A0A0J9TCL7 A0A195DXP8 A0A1A9XXQ7 B4P2G2 B3N7S2 Q9VPR4 H0X1H4 B4Q658 J9JTX5 A0A195FEH4 A0A091DT08 A0A1S3JDQ8 B3MLP8 A0A2K6QQG7 A0A2I2V019 A0A1S3W3K9 F7GHQ8 G3VN49 A0A3Q0DL33 U3DX50 E9ILD0 Q9NVX2 H0VN00 A0A2K6LFA1 A0A287A8X1 M3VUD4 A0A2K5RDR6 G2HGH5 A0A1D5R0W2 B4ICZ0 V9KP55 A0A2K5MYU0 A0A1B0APF9 A0A287ACH7 A0A096P3W2 A0A182VBH9 A0A182HZJ3 Q7PTK2 B2GV82
Pubmed
19121390
26354079
23622113
22118469
28004739
24845553
+ More
23537049 21292972 25348373 24495485 18362917 19820115 20075255 25315136 26580012 26823975 27762356 28648823 9857191 26108605 25474469 20566863 20920257 23761445 17994087 21719571 16305752 19393038 22936249 17550304 10731132 12537572 12537569 25362486 17975172 21709235 25243066 21282665 14702039 16625196 15489334 11124703 12429849 17525332 19413330 21269460 22223895 22814378 26601951 16959974 21993624 16136131 21484476 17431167 25319552 24402279 12364791 14747013 17210077 15057822
23537049 21292972 25348373 24495485 18362917 19820115 20075255 25315136 26580012 26823975 27762356 28648823 9857191 26108605 25474469 20566863 20920257 23761445 17994087 21719571 16305752 19393038 22936249 17550304 10731132 12537572 12537569 25362486 17975172 21709235 25243066 21282665 14702039 16625196 15489334 11124703 12429849 17525332 19413330 21269460 22223895 22814378 26601951 16959974 21993624 16136131 21484476 17431167 25319552 24402279 12364791 14747013 17210077 15057822
EMBL
BABH01013963
ODYU01000113
SOQ34336.1
GDQN01006160
JAT84894.1
RSAL01000041
+ More
RVE50866.1 NWSH01003775 PCG65828.1 KQ459595 KPI95607.1 KQ461154 KPJ08536.1 GAIX01013846 JAA78714.1 AGBW02011442 OWR46648.1 LJIG01015965 KRT82020.1 GEZM01102989 JAV51638.1 KK853689 KDQ97268.1 NEVH01009378 PNF33341.1 GEDC01022265 GEDC01021364 JAS15033.1 JAS15934.1 KB631581 ERL84480.1 GDIP01191686 JAJ31716.1 GDIQ01087187 JAN07550.1 GDIP01146187 JAJ77215.1 GDHF01032634 JAI19680.1 GDIP01222889 JAJ00513.1 GDIP01045716 JAM57999.1 GL732637 EFX69411.1 GDIQ01090749 JAN03988.1 GAKP01006747 GAKP01006745 JAC52205.1 GDHF01002402 JAI49912.1 GECZ01009101 JAS60668.1 GAMC01011422 JAB95133.1 KQ971382 EEZ97691.1 KQ435210 KZC15080.1 KA648474 AFP63103.1 KT984270 ALR88667.1 GECL01001832 JAP04292.1 GDHC01008545 GDHC01007399 JAQ10084.1 JAQ11230.1 CM004469 OCT92242.1 KQ435762 KOX75548.1 NNAY01001258 OXU24594.1 AF069737 BC044710 AAC62236.1 AAH44710.1 GGFJ01004849 MBW53990.1 JRES01001179 KNC24760.1 CCAG010000229 GBBI01002463 JAC16249.1 DS235206 EEB13311.1 BC122040 AAI22041.1 KQ977793 KYM99658.1 ADMH02000120 ETN67716.1 GBYB01015346 JAG85113.1 GGFJ01004848 MBW53989.1 CH933807 EDW13082.2 GL888480 EGI60282.1 BT021777 AGTP01050539 AQIB01146960 CM002910 KMY87340.1 KQ980107 KYN17648.1 CM000157 EDW87158.2 CH954177 EDV57248.1 AE014134 AY089286 AJ012588 AAF51479.2 AAL90024.1 CAA10070.1 AAQR03133423 AAQR03133424 CM000361 EDX03242.1 ABLF02031542 KQ981625 KYN39080.1 KN123290 KFO25941.1 CH902620 EDV30769.1 AANG04004035 AEFK01177033 AEFK01177034 GAMQ01005039 JAB36812.1 GL764074 EFZ18631.1 AK001320 AB209111 AC022916 BC002884 BC012075 AJ005257 AAKN02033115 AEMK02000080 AACZ04043901 AK305839 NBAG03000289 BAK62833.1 PNI47334.1 JSUE03016471 JSUE03016472 JU476081 AFH32885.1 CH480829 EDW45416.1 JW867903 AFP00421.1 JXJN01001399 AHZZ02010044 APCN01001981 AAAB01008799 EAA03804.5 AC095947 BC166567 AAI66567.1
RVE50866.1 NWSH01003775 PCG65828.1 KQ459595 KPI95607.1 KQ461154 KPJ08536.1 GAIX01013846 JAA78714.1 AGBW02011442 OWR46648.1 LJIG01015965 KRT82020.1 GEZM01102989 JAV51638.1 KK853689 KDQ97268.1 NEVH01009378 PNF33341.1 GEDC01022265 GEDC01021364 JAS15033.1 JAS15934.1 KB631581 ERL84480.1 GDIP01191686 JAJ31716.1 GDIQ01087187 JAN07550.1 GDIP01146187 JAJ77215.1 GDHF01032634 JAI19680.1 GDIP01222889 JAJ00513.1 GDIP01045716 JAM57999.1 GL732637 EFX69411.1 GDIQ01090749 JAN03988.1 GAKP01006747 GAKP01006745 JAC52205.1 GDHF01002402 JAI49912.1 GECZ01009101 JAS60668.1 GAMC01011422 JAB95133.1 KQ971382 EEZ97691.1 KQ435210 KZC15080.1 KA648474 AFP63103.1 KT984270 ALR88667.1 GECL01001832 JAP04292.1 GDHC01008545 GDHC01007399 JAQ10084.1 JAQ11230.1 CM004469 OCT92242.1 KQ435762 KOX75548.1 NNAY01001258 OXU24594.1 AF069737 BC044710 AAC62236.1 AAH44710.1 GGFJ01004849 MBW53990.1 JRES01001179 KNC24760.1 CCAG010000229 GBBI01002463 JAC16249.1 DS235206 EEB13311.1 BC122040 AAI22041.1 KQ977793 KYM99658.1 ADMH02000120 ETN67716.1 GBYB01015346 JAG85113.1 GGFJ01004848 MBW53989.1 CH933807 EDW13082.2 GL888480 EGI60282.1 BT021777 AGTP01050539 AQIB01146960 CM002910 KMY87340.1 KQ980107 KYN17648.1 CM000157 EDW87158.2 CH954177 EDV57248.1 AE014134 AY089286 AJ012588 AAF51479.2 AAL90024.1 CAA10070.1 AAQR03133423 AAQR03133424 CM000361 EDX03242.1 ABLF02031542 KQ981625 KYN39080.1 KN123290 KFO25941.1 CH902620 EDV30769.1 AANG04004035 AEFK01177033 AEFK01177034 GAMQ01005039 JAB36812.1 GL764074 EFZ18631.1 AK001320 AB209111 AC022916 BC002884 BC012075 AJ005257 AAKN02033115 AEMK02000080 AACZ04043901 AK305839 NBAG03000289 BAK62833.1 PNI47334.1 JSUE03016471 JSUE03016472 JU476081 AFH32885.1 CH480829 EDW45416.1 JW867903 AFP00421.1 JXJN01001399 AHZZ02010044 APCN01001981 AAAB01008799 EAA03804.5 AC095947 BC166567 AAI66567.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000192223 UP000027135 UP000235965 UP000030742 UP000000305 UP000007266 UP000076502 UP000002358 UP000095301 UP000079169 UP000069272 UP000186698 UP000053105 UP000215335 UP000078200 UP000095300 UP000037069 UP000092444 UP000009046 UP000078542 UP000000673 UP000009192 UP000007755 UP000091820 UP000009136 UP000005215 UP000029965 UP000078492 UP000092443 UP000002282 UP000008711 UP000000803 UP000005225 UP000000304 UP000007819 UP000078541 UP000028990 UP000085678 UP000007801 UP000233200 UP000011712 UP000079721 UP000008225 UP000007648 UP000189704 UP000005640 UP000005447 UP000233180 UP000008227 UP000233040 UP000002277 UP000006718 UP000001292 UP000233060 UP000092460 UP000028761 UP000075903 UP000075840 UP000007062 UP000002494
UP000192223 UP000027135 UP000235965 UP000030742 UP000000305 UP000007266 UP000076502 UP000002358 UP000095301 UP000079169 UP000069272 UP000186698 UP000053105 UP000215335 UP000078200 UP000095300 UP000037069 UP000092444 UP000009046 UP000078542 UP000000673 UP000009192 UP000007755 UP000091820 UP000009136 UP000005215 UP000029965 UP000078492 UP000092443 UP000002282 UP000008711 UP000000803 UP000005225 UP000000304 UP000007819 UP000078541 UP000028990 UP000085678 UP000007801 UP000233200 UP000011712 UP000079721 UP000008225 UP000007648 UP000189704 UP000005640 UP000005447 UP000233180 UP000008227 UP000233040 UP000002277 UP000006718 UP000001292 UP000233060 UP000092460 UP000028761 UP000075903 UP000075840 UP000007062 UP000002494
Interpro
IPR017986
WD40_repeat_dom
+ More
IPR001680 WD40_repeat
IPR001632 Gprotein_B
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR020472 G-protein_beta_WD-40_rep
IPR012972 NLE
IPR011047 Quinoprotein_ADH-like_supfam
IPR001320 Iontro_rcpt
IPR024977 Apc4_WD40_dom
IPR011041 Quinoprot_gluc/sorb_DH
IPR001680 WD40_repeat
IPR001632 Gprotein_B
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR020472 G-protein_beta_WD-40_rep
IPR012972 NLE
IPR011047 Quinoprotein_ADH-like_supfam
IPR001320 Iontro_rcpt
IPR024977 Apc4_WD40_dom
IPR011041 Quinoprot_gluc/sorb_DH
Gene 3D
ProteinModelPortal
H9J720
A0A2H1V0Q1
A0A1E1WD31
A0A3S2TNH8
A0A2A4J2B7
A0A194PQM6
+ More
A0A194QY83 S4NNP5 A0A212EYS4 A0A0T6B410 A0A1Y1JU29 A0A1W4WF11 A0A067QEW1 A0A2J7QXN3 A0A1B6CNS1 U4U3B9 A0A0P5B0P2 A0A0P6CUL8 A0A0P5E6A5 A0A0K8U036 A0A0N7ZQD1 A0A0P5ZCM1 E9HFT7 A0A0P6CI94 A0A034WDM4 A0A0K8WFQ0 A0A1B6GE01 W8BDS3 D6X557 A0A154PV51 K7J2R3 A0A1I8NEG9 A0A1S3D021 T1PIJ4 A0A0U2K782 A0A0V0G8D6 A0A146LXZ6 A0A182FB76 A0A1L8H809 A0A0N0BH53 A0A232F1K0 Q7ZXK9 A0A1A9VBQ2 A0A2M4BLN3 A0A1I8NN00 A0A0L0BXN3 A0A1B0G8B6 A0A023F589 E0VIV5 Q0D2B0 A0A151IFH4 W5JU99 A0A0C9S352 A0A2M4BLM9 B4KFG7 F4WZK7 A0A1A9WN04 Q58D20 A0A3Q1N213 I3M7E4 A0A0D9R1W1 A0A0J9TCL7 A0A195DXP8 A0A1A9XXQ7 B4P2G2 B3N7S2 Q9VPR4 H0X1H4 B4Q658 J9JTX5 A0A195FEH4 A0A091DT08 A0A1S3JDQ8 B3MLP8 A0A2K6QQG7 A0A2I2V019 A0A1S3W3K9 F7GHQ8 G3VN49 A0A3Q0DL33 U3DX50 E9ILD0 Q9NVX2 H0VN00 A0A2K6LFA1 A0A287A8X1 M3VUD4 A0A2K5RDR6 G2HGH5 A0A1D5R0W2 B4ICZ0 V9KP55 A0A2K5MYU0 A0A1B0APF9 A0A287ACH7 A0A096P3W2 A0A182VBH9 A0A182HZJ3 Q7PTK2 B2GV82
A0A194QY83 S4NNP5 A0A212EYS4 A0A0T6B410 A0A1Y1JU29 A0A1W4WF11 A0A067QEW1 A0A2J7QXN3 A0A1B6CNS1 U4U3B9 A0A0P5B0P2 A0A0P6CUL8 A0A0P5E6A5 A0A0K8U036 A0A0N7ZQD1 A0A0P5ZCM1 E9HFT7 A0A0P6CI94 A0A034WDM4 A0A0K8WFQ0 A0A1B6GE01 W8BDS3 D6X557 A0A154PV51 K7J2R3 A0A1I8NEG9 A0A1S3D021 T1PIJ4 A0A0U2K782 A0A0V0G8D6 A0A146LXZ6 A0A182FB76 A0A1L8H809 A0A0N0BH53 A0A232F1K0 Q7ZXK9 A0A1A9VBQ2 A0A2M4BLN3 A0A1I8NN00 A0A0L0BXN3 A0A1B0G8B6 A0A023F589 E0VIV5 Q0D2B0 A0A151IFH4 W5JU99 A0A0C9S352 A0A2M4BLM9 B4KFG7 F4WZK7 A0A1A9WN04 Q58D20 A0A3Q1N213 I3M7E4 A0A0D9R1W1 A0A0J9TCL7 A0A195DXP8 A0A1A9XXQ7 B4P2G2 B3N7S2 Q9VPR4 H0X1H4 B4Q658 J9JTX5 A0A195FEH4 A0A091DT08 A0A1S3JDQ8 B3MLP8 A0A2K6QQG7 A0A2I2V019 A0A1S3W3K9 F7GHQ8 G3VN49 A0A3Q0DL33 U3DX50 E9ILD0 Q9NVX2 H0VN00 A0A2K6LFA1 A0A287A8X1 M3VUD4 A0A2K5RDR6 G2HGH5 A0A1D5R0W2 B4ICZ0 V9KP55 A0A2K5MYU0 A0A1B0APF9 A0A287ACH7 A0A096P3W2 A0A182VBH9 A0A182HZJ3 Q7PTK2 B2GV82
PDB
6FT6
E-value=6.30214e-121,
Score=1112
Ontologies
GO
GO:0005730
GO:0003743
GO:0042254
GO:0016021
GO:0004970
GO:0000027
GO:0007219
GO:0003824
GO:0045746
GO:0005112
GO:0045747
GO:0090263
GO:2001268
GO:0001826
GO:0042273
GO:0048705
GO:0061484
GO:0045930
GO:0001756
GO:0001822
GO:0005634
GO:0005515
GO:0005509
GO:0006468
GO:0007165
GO:0005085
GO:0005089
GO:0035023
GO:0016742
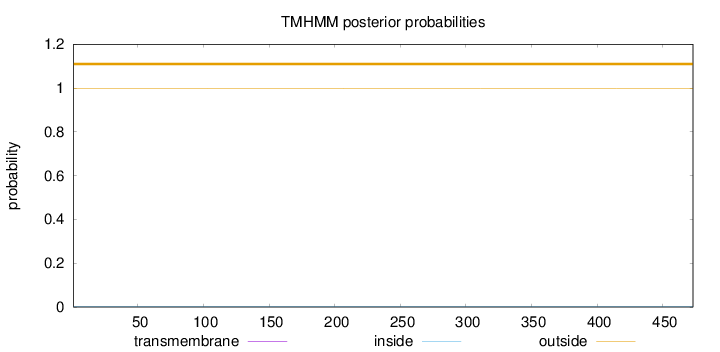
Topology
Subcellular location
Length:
473
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00243
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00251
outside
1 - 473
Population Genetic Test Statistics
Pi
190.568241
Theta
196.655772
Tajima's D
-0.30665
CLR
2.564053
CSRT
0.294685265736713
Interpretation
Uncertain
