Pre Gene Modal
BGIBMGA005498
Annotation
PREDICTED:_something_about_silencing_protein_10-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 4.572
Sequence
CDS
ATGGCTGATAAAATAAGAATAAATTCAAGATTTGACCAAAAAGAGAATTATGCCCCTTCAGATTCAGAAGATGGTTATTCTGAAAATGAAAAGCGATTATTGGATAAAGTACGTAAGAGAAAACATGAGTCTGATAGTGAAGAGGAAATGTATGCTTTCTCTGATTCTGACGAGGAATCAGATCAAAAGGATGATTTGGGAATAGCAGACAGTGATGTTGAAGGACAGGAAAAATCAGACGATGACTTACCTGATTCAAAAGCTTGGGGCAAAAATAAACAATCATACTATGCCACTGATTATGTTGACAAAGATTATGGAGGCTTCGGTGATGATGAAGAAGCTGCTCTTATGGAAGAAGAGGAGGCTAAGAATATTCAGAAGAGATTGCTAGAACAACTTGGAGATGAAGATTTTACCTTGGATTTCTTTACTAAACATGTACAAGACGCTGAAGAAAGAGAAACAGTAATAAAAAGTGATTTGAGCCAGTTGTCAAAAAGACAAAAGCTACAATTGTTTGAAAAGGAAAGTCCTGAGTTTTCAGGTCTTGTTGATGACTTCCAAACAAAACTTACTGTAGCTAAAGATAACCTACATCCAGTCTTGGAATTAGTAAAAGATGGCAAATTACCAAAATGTGCAGCATCAAAGTTTGTAAAAACAAACTATGATCTAATTCTAAATTATTGTACAAATATTAGTTTTTATTTGCTTCTAAAGAGTCAGCGAGTAAACATTCAAAATCATCCAGTTATAAAAAGGTTGTATCAGTACAGACAAATGTTGAATAAAATGGAACCGATATATTTGGAAGTAATTAAACCGCAAATAGATAAAATTCTTCTGGCTGTGAAGAACAACCTTGTTTTGGAAGTAAATGAACAAAAAGAAGCTAAGAAACGCAAGTTAGAAAAACTTATTTCAGAATCAACTCCCAAGAAATTGAAACTTATCAGTGCAATAGAAAAAGATGAAGGAGAAGACACTGGAGTATCTGATGATGAAGAATATGAAGGGAATGATTACTTTAAAGATAACGATAAAGTTGAAAATGTGCCAAGAAGTAATGAATCTGAAGAAAGTGGTTTTTCAGATAATGAAGATACTGTACTTGGTGAAACAGAAGTTGACCATAATAAACCATCAACATCATCTCAACAAATTGTTACAGAAGATCTTGGAATGAAAAGAGAAATAACATATCAAATTGCAAAGAATAAAGGGTTGACACCCCATCGAAAGAAAGATCAAAGAAACCCAAGAGTCAAACATAAACTTAAATACAGAAAGGCTAAAATCAGAAGGAAGGGTGCAGTGAGGGAACCTAAGACAGAAGTGTCCAGATATGGTGGTGAATCATCAGGCATAAAAGCAAATTTGAAAAAGAGCATAACAATCAAGTAA
Protein
MADKIRINSRFDQKENYAPSDSEDGYSENEKRLLDKVRKRKHESDSEEEMYAFSDSDEESDQKDDLGIADSDVEGQEKSDDDLPDSKAWGKNKQSYYATDYVDKDYGGFGDDEEAALMEEEEAKNIQKRLLEQLGDEDFTLDFFTKHVQDAEERETVIKSDLSQLSKRQKLQLFEKESPEFSGLVDDFQTKLTVAKDNLHPVLELVKDGKLPKCAASKFVKTNYDLILNYCTNISFYLLLKSQRVNIQNHPVIKRLYQYRQMLNKMEPIYLEVIKPQIDKILLAVKNNLVLEVNEQKEAKKRKLEKLISESTPKKLKLISAIEKDEGEDTGVSDDEEYEGNDYFKDNDKVENVPRSNESEESGFSDNEDTVLGETEVDHNKPSTSSQQIVTEDLGMKREITYQIAKNKGLTPHRKKDQRNPRVKHKLKYRKAKIRRKGAVREPKTEVSRYGGESSGIKANLKKSITIK
Summary
Uniprot
H9J7K6
A0A1E1VZV1
A0A2H1V0G0
A0A0L7KYY1
A0A194QU72
A0A194PQ85
+ More
A0A212EYX1 D2A199 A0A067QV45 A0A0T6AUD8 A0A2J7QKN1 A0A232EQI2 K7J7M2 A0A0L7RDC9 A0A1B6GCQ4 A0A1B6IZ76 A0A088ART1 A0A2A3E2F0 A0A0M8ZPX1 A0A2J7QKM7 A0A1I8P5T7 A0A1J1J4H9 A0A336MZR0 A0A336L112 W8BSG4 A0A1B6CGF7 A0A1B0G0S7 B7Q4P6 D3TNZ4 A0A0P5WSW8 A0A131XMB8 J9K8I1 A0A0P5WSC6 A0A0K8RBJ7 B4L7T1 A0A0P5B111 A0A1Q3F2W3 A0A0N8CS31 A0A0N8A2K1 A0A0P6GK81 A0A0P5VEF7 A0A3B0KRK4 A0A0P5STV4 A0A0P6FQP2 A0A0P5V7V0 A0A0P5U2Y2 A0A0P5YA89 A0A0K8RCL5 A0A131XZJ2 A0A1Q3F4L2 A0A0N8DBN2 A0A0L0CDW2 A0A1E1XSY4 A0A1E1XB62 E2BX52 A0A210PWI1 A0A3Q3MQ81 A0A146ZW45 A0A3B4UNZ3 A0A3Q1BE58 A0A2R5L8N4 A0A0P5DXE6 W5LZV8 A0A3Q2QW53 A0A0P5CJE3 A0A3B1IT18 G1SYC5 M7BPN6 A0A3P8S1P1 A0A3P9NDY9 A0A3Q1FGR1 A0A1W4XYH1 A0A1S3P2I1 A0A2K5E3C2 A0A2Y9DZ79 A0A3Q2ZRN0 K7EWE5 A0A3B3XNQ3 A0A2K5RKQ3 A0A0P5WTD4 A0A3P9AMF0 A0A3Q2CW15 A0A158NHK4 A0A2K6SEX0 W5LZX7 A0A1S3P289 A0A293MXF1 F6QJZ7 H0X7X6 A0A0N8CYE3 G1KMV7 A0A1S3FII0 W5PD55 A0A1A8JWR1 A0A3B3CSB4 A0A1A8D4D6 K1PNZ4
A0A212EYX1 D2A199 A0A067QV45 A0A0T6AUD8 A0A2J7QKN1 A0A232EQI2 K7J7M2 A0A0L7RDC9 A0A1B6GCQ4 A0A1B6IZ76 A0A088ART1 A0A2A3E2F0 A0A0M8ZPX1 A0A2J7QKM7 A0A1I8P5T7 A0A1J1J4H9 A0A336MZR0 A0A336L112 W8BSG4 A0A1B6CGF7 A0A1B0G0S7 B7Q4P6 D3TNZ4 A0A0P5WSW8 A0A131XMB8 J9K8I1 A0A0P5WSC6 A0A0K8RBJ7 B4L7T1 A0A0P5B111 A0A1Q3F2W3 A0A0N8CS31 A0A0N8A2K1 A0A0P6GK81 A0A0P5VEF7 A0A3B0KRK4 A0A0P5STV4 A0A0P6FQP2 A0A0P5V7V0 A0A0P5U2Y2 A0A0P5YA89 A0A0K8RCL5 A0A131XZJ2 A0A1Q3F4L2 A0A0N8DBN2 A0A0L0CDW2 A0A1E1XSY4 A0A1E1XB62 E2BX52 A0A210PWI1 A0A3Q3MQ81 A0A146ZW45 A0A3B4UNZ3 A0A3Q1BE58 A0A2R5L8N4 A0A0P5DXE6 W5LZV8 A0A3Q2QW53 A0A0P5CJE3 A0A3B1IT18 G1SYC5 M7BPN6 A0A3P8S1P1 A0A3P9NDY9 A0A3Q1FGR1 A0A1W4XYH1 A0A1S3P2I1 A0A2K5E3C2 A0A2Y9DZ79 A0A3Q2ZRN0 K7EWE5 A0A3B3XNQ3 A0A2K5RKQ3 A0A0P5WTD4 A0A3P9AMF0 A0A3Q2CW15 A0A158NHK4 A0A2K6SEX0 W5LZX7 A0A1S3P289 A0A293MXF1 F6QJZ7 H0X7X6 A0A0N8CYE3 G1KMV7 A0A1S3FII0 W5PD55 A0A1A8JWR1 A0A3B3CSB4 A0A1A8D4D6 K1PNZ4
Pubmed
EMBL
BABH01013962
BABH01013963
GDQN01010840
JAT80214.1
ODYU01000113
SOQ34335.1
+ More
JTDY01004221 KOB68437.1 KQ461154 KPJ08535.1 KQ459595 KPI95606.1 AGBW02011442 OWR46647.1 KQ971338 EFA01562.1 KK852902 KDR14064.1 LJIG01022784 KRT78737.1 NEVH01013262 PNF29141.1 NNAY01002764 OXU20615.1 AAZX01002179 KQ414614 KOC68829.1 GECZ01009547 JAS60222.1 GECU01015490 JAS92216.1 KZ288427 PBC25877.1 KQ438551 KOX67169.1 PNF29142.1 CVRI01000070 CRL07304.1 UFQS01002855 UFQT01002855 SSX14771.1 SSX34163.1 UFQS01001439 UFQT01001439 SSX10906.1 SSX30584.1 GAMC01004475 GAMC01004473 JAC02083.1 GEDC01024883 JAS12415.1 CCAG010004515 ABJB010027122 ABJB010110019 ABJB010371168 ABJB010666651 ABJB011072904 DS857337 EEC13817.1 EZ423146 ADD19422.1 GDIP01082088 JAM21627.1 GEFH01001715 JAP66866.1 ABLF02035529 GDIP01082090 JAM21625.1 GADI01005595 JAA68213.1 CH933814 EDW05506.1 GDIP01191331 JAJ32071.1 GFDL01013151 JAV21894.1 GDIP01104399 JAL99315.1 GDIP01188729 JAJ34673.1 GDIQ01042563 JAN52174.1 GDIP01101446 JAM02269.1 OUUW01000019 SPP89279.1 GDIP01191332 GDIP01135353 LRGB01003146 JAL68361.1 KZS04022.1 GDIQ01055656 JAN39081.1 GDIP01104400 JAL99314.1 GDIP01118898 JAL84816.1 GDIP01060648 JAM43067.1 GADI01005594 JAA68214.1 GEFM01003143 JAP72653.1 GFDL01012539 JAV22506.1 GDIP01049591 JAM54124.1 JRES01000523 KNC30435.1 GFAA01001016 JAU02419.1 GFAC01002708 JAT96480.1 GL451202 EFN79771.1 NEDP02005443 OWF40834.1 GCES01015782 JAR70541.1 GGLE01001651 MBY05777.1 GDIP01151192 JAJ72210.1 AHAT01034526 AHAT01034527 AHAT01034528 AHAT01034529 AHAT01034530 AHAT01034531 AHAT01034532 AHAT01034533 AHAT01034534 AHAT01034535 GDIP01169626 JAJ53776.1 AAGW02023094 KB551681 EMP30137.1 AGCU01157539 GDIP01082089 JAM21626.1 ADTU01015877 GFWV01019860 MAA44588.1 AAQR03168010 GDIP01086703 GDIQ01055657 JAM17012.1 JAN39080.1 AMGL01099452 HAEE01003804 SBR23824.1 HADZ01022152 HAEA01002906 SBP86093.1 JH815775 EKC18200.1
JTDY01004221 KOB68437.1 KQ461154 KPJ08535.1 KQ459595 KPI95606.1 AGBW02011442 OWR46647.1 KQ971338 EFA01562.1 KK852902 KDR14064.1 LJIG01022784 KRT78737.1 NEVH01013262 PNF29141.1 NNAY01002764 OXU20615.1 AAZX01002179 KQ414614 KOC68829.1 GECZ01009547 JAS60222.1 GECU01015490 JAS92216.1 KZ288427 PBC25877.1 KQ438551 KOX67169.1 PNF29142.1 CVRI01000070 CRL07304.1 UFQS01002855 UFQT01002855 SSX14771.1 SSX34163.1 UFQS01001439 UFQT01001439 SSX10906.1 SSX30584.1 GAMC01004475 GAMC01004473 JAC02083.1 GEDC01024883 JAS12415.1 CCAG010004515 ABJB010027122 ABJB010110019 ABJB010371168 ABJB010666651 ABJB011072904 DS857337 EEC13817.1 EZ423146 ADD19422.1 GDIP01082088 JAM21627.1 GEFH01001715 JAP66866.1 ABLF02035529 GDIP01082090 JAM21625.1 GADI01005595 JAA68213.1 CH933814 EDW05506.1 GDIP01191331 JAJ32071.1 GFDL01013151 JAV21894.1 GDIP01104399 JAL99315.1 GDIP01188729 JAJ34673.1 GDIQ01042563 JAN52174.1 GDIP01101446 JAM02269.1 OUUW01000019 SPP89279.1 GDIP01191332 GDIP01135353 LRGB01003146 JAL68361.1 KZS04022.1 GDIQ01055656 JAN39081.1 GDIP01104400 JAL99314.1 GDIP01118898 JAL84816.1 GDIP01060648 JAM43067.1 GADI01005594 JAA68214.1 GEFM01003143 JAP72653.1 GFDL01012539 JAV22506.1 GDIP01049591 JAM54124.1 JRES01000523 KNC30435.1 GFAA01001016 JAU02419.1 GFAC01002708 JAT96480.1 GL451202 EFN79771.1 NEDP02005443 OWF40834.1 GCES01015782 JAR70541.1 GGLE01001651 MBY05777.1 GDIP01151192 JAJ72210.1 AHAT01034526 AHAT01034527 AHAT01034528 AHAT01034529 AHAT01034530 AHAT01034531 AHAT01034532 AHAT01034533 AHAT01034534 AHAT01034535 GDIP01169626 JAJ53776.1 AAGW02023094 KB551681 EMP30137.1 AGCU01157539 GDIP01082089 JAM21626.1 ADTU01015877 GFWV01019860 MAA44588.1 AAQR03168010 GDIP01086703 GDIQ01055657 JAM17012.1 JAN39080.1 AMGL01099452 HAEE01003804 SBR23824.1 HADZ01022152 HAEA01002906 SBP86093.1 JH815775 EKC18200.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000053268
UP000007151
UP000007266
+ More
UP000027135 UP000235965 UP000215335 UP000002358 UP000053825 UP000005203 UP000242457 UP000053105 UP000095300 UP000183832 UP000092444 UP000001555 UP000007819 UP000009192 UP000268350 UP000076858 UP000037069 UP000008237 UP000242188 UP000261640 UP000261420 UP000257160 UP000018468 UP000265000 UP000018467 UP000001811 UP000031443 UP000265080 UP000242638 UP000257200 UP000192224 UP000087266 UP000233020 UP000248480 UP000264800 UP000007267 UP000261480 UP000233040 UP000265140 UP000265020 UP000005205 UP000233220 UP000002281 UP000005225 UP000001646 UP000081671 UP000002356 UP000261560 UP000005408
UP000027135 UP000235965 UP000215335 UP000002358 UP000053825 UP000005203 UP000242457 UP000053105 UP000095300 UP000183832 UP000092444 UP000001555 UP000007819 UP000009192 UP000268350 UP000076858 UP000037069 UP000008237 UP000242188 UP000261640 UP000261420 UP000257160 UP000018468 UP000265000 UP000018467 UP000001811 UP000031443 UP000265080 UP000242638 UP000257200 UP000192224 UP000087266 UP000233020 UP000248480 UP000264800 UP000007267 UP000261480 UP000233040 UP000265140 UP000265020 UP000005205 UP000233220 UP000002281 UP000005225 UP000001646 UP000081671 UP000002356 UP000261560 UP000005408
PRIDE
ProteinModelPortal
H9J7K6
A0A1E1VZV1
A0A2H1V0G0
A0A0L7KYY1
A0A194QU72
A0A194PQ85
+ More
A0A212EYX1 D2A199 A0A067QV45 A0A0T6AUD8 A0A2J7QKN1 A0A232EQI2 K7J7M2 A0A0L7RDC9 A0A1B6GCQ4 A0A1B6IZ76 A0A088ART1 A0A2A3E2F0 A0A0M8ZPX1 A0A2J7QKM7 A0A1I8P5T7 A0A1J1J4H9 A0A336MZR0 A0A336L112 W8BSG4 A0A1B6CGF7 A0A1B0G0S7 B7Q4P6 D3TNZ4 A0A0P5WSW8 A0A131XMB8 J9K8I1 A0A0P5WSC6 A0A0K8RBJ7 B4L7T1 A0A0P5B111 A0A1Q3F2W3 A0A0N8CS31 A0A0N8A2K1 A0A0P6GK81 A0A0P5VEF7 A0A3B0KRK4 A0A0P5STV4 A0A0P6FQP2 A0A0P5V7V0 A0A0P5U2Y2 A0A0P5YA89 A0A0K8RCL5 A0A131XZJ2 A0A1Q3F4L2 A0A0N8DBN2 A0A0L0CDW2 A0A1E1XSY4 A0A1E1XB62 E2BX52 A0A210PWI1 A0A3Q3MQ81 A0A146ZW45 A0A3B4UNZ3 A0A3Q1BE58 A0A2R5L8N4 A0A0P5DXE6 W5LZV8 A0A3Q2QW53 A0A0P5CJE3 A0A3B1IT18 G1SYC5 M7BPN6 A0A3P8S1P1 A0A3P9NDY9 A0A3Q1FGR1 A0A1W4XYH1 A0A1S3P2I1 A0A2K5E3C2 A0A2Y9DZ79 A0A3Q2ZRN0 K7EWE5 A0A3B3XNQ3 A0A2K5RKQ3 A0A0P5WTD4 A0A3P9AMF0 A0A3Q2CW15 A0A158NHK4 A0A2K6SEX0 W5LZX7 A0A1S3P289 A0A293MXF1 F6QJZ7 H0X7X6 A0A0N8CYE3 G1KMV7 A0A1S3FII0 W5PD55 A0A1A8JWR1 A0A3B3CSB4 A0A1A8D4D6 K1PNZ4
A0A212EYX1 D2A199 A0A067QV45 A0A0T6AUD8 A0A2J7QKN1 A0A232EQI2 K7J7M2 A0A0L7RDC9 A0A1B6GCQ4 A0A1B6IZ76 A0A088ART1 A0A2A3E2F0 A0A0M8ZPX1 A0A2J7QKM7 A0A1I8P5T7 A0A1J1J4H9 A0A336MZR0 A0A336L112 W8BSG4 A0A1B6CGF7 A0A1B0G0S7 B7Q4P6 D3TNZ4 A0A0P5WSW8 A0A131XMB8 J9K8I1 A0A0P5WSC6 A0A0K8RBJ7 B4L7T1 A0A0P5B111 A0A1Q3F2W3 A0A0N8CS31 A0A0N8A2K1 A0A0P6GK81 A0A0P5VEF7 A0A3B0KRK4 A0A0P5STV4 A0A0P6FQP2 A0A0P5V7V0 A0A0P5U2Y2 A0A0P5YA89 A0A0K8RCL5 A0A131XZJ2 A0A1Q3F4L2 A0A0N8DBN2 A0A0L0CDW2 A0A1E1XSY4 A0A1E1XB62 E2BX52 A0A210PWI1 A0A3Q3MQ81 A0A146ZW45 A0A3B4UNZ3 A0A3Q1BE58 A0A2R5L8N4 A0A0P5DXE6 W5LZV8 A0A3Q2QW53 A0A0P5CJE3 A0A3B1IT18 G1SYC5 M7BPN6 A0A3P8S1P1 A0A3P9NDY9 A0A3Q1FGR1 A0A1W4XYH1 A0A1S3P2I1 A0A2K5E3C2 A0A2Y9DZ79 A0A3Q2ZRN0 K7EWE5 A0A3B3XNQ3 A0A2K5RKQ3 A0A0P5WTD4 A0A3P9AMF0 A0A3Q2CW15 A0A158NHK4 A0A2K6SEX0 W5LZX7 A0A1S3P289 A0A293MXF1 F6QJZ7 H0X7X6 A0A0N8CYE3 G1KMV7 A0A1S3FII0 W5PD55 A0A1A8JWR1 A0A3B3CSB4 A0A1A8D4D6 K1PNZ4
PDB
5OQL
E-value=1.20034e-08,
Score=143
Ontologies
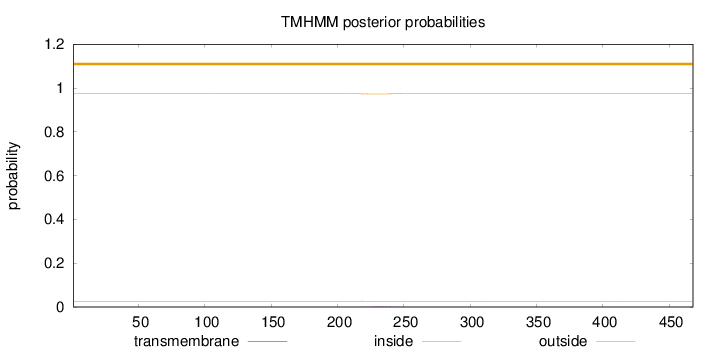
Topology
Length:
468
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03942
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02664
outside
1 - 468
Population Genetic Test Statistics
Pi
160.041339
Theta
186.503217
Tajima's D
-0.505629
CLR
0.43558
CSRT
0.23908804559772
Interpretation
Uncertain
