Pre Gene Modal
BGIBMGA005496
Annotation
cytochrome_P450?_family_315?_subfamily_a?_polypeptide_1_[Bombyx_mori]
Full name
Cytochrome P450 315a1, mitochondrial
Alternative Name
CYPCCCXVA1
Protein shadow
Protein shadow
Location in the cell
Mitochondrial Reliability : 2.578
Sequence
CDS
ATGCATCGTTTCCCGTCGATGTCTTCCATTAGATCCGCTGTTAGGAGTCGTAACAGTAATCGATGTTCAATGTCAACTAAGCCTCATAAATCTCTACGTACCATAGATGAGATGCCACATAAGAAATCATTACCAATCATAGGAACAAAATTTGACTTATTTTCTGCAGGTGGAGGAAAAAACTTACACAAATATATCGACATGCGTCACAAGCAATTAGGACCTATATTCTACGAGAGATTAACAGGAAAGACAAAACTTGTCTTTATCAGCGACCCAACTCACATGAAAAGTTTATTCTTAAATTTAGAAGGAAAATATCCGGCACATATTTTACCAGAACCGTGGGTACTATATGAAAAATTATACGGATCAAAAAGAGGACTTTTCTTTATGGACGGAGAGGATTGGTTGATTAATCGACGAATTATGAACAAACATCTACTCAGAGAAGATTCAGATGTATGGTTACGAGCTCCTATAAGAACAGCTGTGTTTCACTTTATATGTAACTGGAAACTTAGAGCTCAAAGTGGCAACTTCTCTCCTAACTTGGAATCTGAGTTTTATAGATTTTCCACTGACGTTATTTTAGCAGTTCTTCAGGGGAATAGTGCACTATTAAAGCCCACGCCAGAATACGAGATGTTATTGTTACTGTTTTCTGAGGCTGTCAAGAAAATATTCTCAACAACTACAAAATTATATGCTTTACCCGTAGAATTCTGCCAACGATGGAATTTAAAAGTGTGGAGAAATTTCAAACAATCCGTTGATGATTCCATCTCTATCGCTCAGAAAATTGTATACGAAATGTTGCACACAAAAGACGCTGGTGACGGTTTGGTAAAAAGACTAAAGGATGAAAATATGTCTGATGAATTAATAACAAGAATTGTTGCTGACTTTGTCATCGCCGCTGGAGATACGACAGCTTATACATCACTTTGGATATTATTTTTACTTTCAAATAATACTGAAATCTTAACTGAAATGAACGACAACGACCAATACGTGAAAAATGTTGTTAAAGAGGCAATGAGATTATATCCTGTAGCACCATTTCTTACAAGAATTTTACCCAAACAATGTGTTTTGGGACCTTATTTGCTCGAGGAAGGGACTCCAGTAATAGCTTCAATCTATACATCGGGTCGAGACGAACAAAATTTTAGCAAAGCAGACCAATTTTTGCCTTATCGATGGGACAGGAACGATCAACGAAAGAAGGATTTAGTAAATCATGTACCATCTGCCACACTGCCATTTGCCTTCGGCGCCAGATCATGTATCGGTAAAAAGATGGCTATGCTACAAATGACAGAATTAATCAGTCAGATTGTGAAAAATTTTGACCTGAAATCTATGAACAATTCTGATGTGGATGCTGTAACTTCACAGGTGTTAGTGCCGAACAAAGACATCAAAGTTTTAATACTACCGCGTTCAATTTCTAAGTAA
Protein
MHRFPSMSSIRSAVRSRNSNRCSMSTKPHKSLRTIDEMPHKKSLPIIGTKFDLFSAGGGKNLHKYIDMRHKQLGPIFYERLTGKTKLVFISDPTHMKSLFLNLEGKYPAHILPEPWVLYEKLYGSKRGLFFMDGEDWLINRRIMNKHLLREDSDVWLRAPIRTAVFHFICNWKLRAQSGNFSPNLESEFYRFSTDVILAVLQGNSALLKPTPEYEMLLLLFSEAVKKIFSTTTKLYALPVEFCQRWNLKVWRNFKQSVDDSISIAQKIVYEMLHTKDAGDGLVKRLKDENMSDELITRIVADFVIAAGDTTAYTSLWILFLLSNNTEILTEMNDNDQYVKNVVKEAMRLYPVAPFLTRILPKQCVLGPYLLEEGTPVIASIYTSGRDEQNFSKADQFLPYRWDRNDQRKKDLVNHVPSATLPFAFGARSCIGKKMAMLQMTELISQIVKNFDLKSMNNSDVDAVTSQVLVPNKDIKVLILPRSISK
Summary
Description
Required for CNS development: midline glial cells. Involved in the metabolism of insect hormones: responsible for ecdysteroid C2-hydroxylase activity. May be involved in the breakdown of synthetic insecticides.
Cofactor
heme
Miscellaneous
Member of the Halloween gene group.
Similarity
Belongs to the cytochrome P450 family.
Belongs to the OXA1/ALB3/YidC family.
Belongs to the OXA1/ALB3/YidC family.
Keywords
Complete proteome
Heme
Iron
Membrane
Metal-binding
Mitochondrion
Monooxygenase
Oxidoreductase
Reference proteome
Transit peptide
Feature
chain Cytochrome P450 315a1, mitochondrial
Uniprot
Q6I6M8
L0N6K4
L0N747
L0N7B2
Q2HZZ4
A0A0P0HKS9
+ More
S6B478 A0A3S2NLQ3 D1GEJ6 A0A248QEJ3 A0A2H1VI64 A0A0L7LLN4 A0A067RDD3 A0A1Y1MMR8 A0A0T6AXK2 D6WKZ9 A0A2J7PGM2 V5K5L7 A0A1W6L1B8 A0A1A9V720 A0A1A9ZL34 B4NK58 A0A1I8N6Z5 A0A034VQK8 A0A088APZ6 A0A0A1X8D5 A0A0L0CBJ1 A0A0L7R8B6 T1HQU7 A0A232F4V0 A0A0K8U898 A0A1I8Q149 B4PTF1 A0A3B0KBX2 A0A2A3EJT6 A0A224XQA4 A0A195C616 Q295B9 Q16NV1 B4GMB6 A0A023F0S2 A0A0P6IXI5 A0A075BFL1 B4HHW3 A0A146M3K4 A0A1B6CUW2 B3P4E3 Q563C5 Q9VGH1 A0A0M4EJV6 B4QTK3 K7IRZ9 A0A1B0CLM4 A0A2H8TGC2 E0VMW9 A0A146LLE3 A0A026W166 A0A1W4WC86 B3MTT6 A0A1A9X2R5 T1JAE9 A0A034VPJ7 B4K9Z0 B4JG07 A0A151WSX3 A0A1A9XW33 A0A1B0BDP7 A0A182N8F3 A0A1J1IJC8 A0A195E966 A0A1B6FRJ4 A0A158NF50 X1WI94 A0A182YH61 B4LXY5 E9ID25 T1J337 E2B079 A0A195FL49 A0A195AUU7 A0A087TZR9 A0A182RHK3 A9Q617 F4WEI7 A0A182MK14 A0A182U3F5 Q7QF34 A0A224XHL4 A0A2S2QD38 A0A0K0LB69 A0A0J7KK56 A0A182QLY6 A0A182KCS5 A0A182VHA7
S6B478 A0A3S2NLQ3 D1GEJ6 A0A248QEJ3 A0A2H1VI64 A0A0L7LLN4 A0A067RDD3 A0A1Y1MMR8 A0A0T6AXK2 D6WKZ9 A0A2J7PGM2 V5K5L7 A0A1W6L1B8 A0A1A9V720 A0A1A9ZL34 B4NK58 A0A1I8N6Z5 A0A034VQK8 A0A088APZ6 A0A0A1X8D5 A0A0L0CBJ1 A0A0L7R8B6 T1HQU7 A0A232F4V0 A0A0K8U898 A0A1I8Q149 B4PTF1 A0A3B0KBX2 A0A2A3EJT6 A0A224XQA4 A0A195C616 Q295B9 Q16NV1 B4GMB6 A0A023F0S2 A0A0P6IXI5 A0A075BFL1 B4HHW3 A0A146M3K4 A0A1B6CUW2 B3P4E3 Q563C5 Q9VGH1 A0A0M4EJV6 B4QTK3 K7IRZ9 A0A1B0CLM4 A0A2H8TGC2 E0VMW9 A0A146LLE3 A0A026W166 A0A1W4WC86 B3MTT6 A0A1A9X2R5 T1JAE9 A0A034VPJ7 B4K9Z0 B4JG07 A0A151WSX3 A0A1A9XW33 A0A1B0BDP7 A0A182N8F3 A0A1J1IJC8 A0A195E966 A0A1B6FRJ4 A0A158NF50 X1WI94 A0A182YH61 B4LXY5 E9ID25 T1J337 E2B079 A0A195FL49 A0A195AUU7 A0A087TZR9 A0A182RHK3 A9Q617 F4WEI7 A0A182MK14 A0A182U3F5 Q7QF34 A0A224XHL4 A0A2S2QD38 A0A0K0LB69 A0A0J7KK56 A0A182QLY6 A0A182KCS5 A0A182VHA7
EC Number
1.14.99.-
Pubmed
15197185
16503480
19682519
28881445
26227816
24845553
+ More
28004739 18362917 19820115 24267698 17994087 25315136 25348373 25830018 26108605 28648823 17550304 15632085 17510324 25474469 26999592 26823975 15804580 12177427 10731132 12537572 14610274 20075255 20566863 24508170 21347285 25244985 21282665 20798317 19060216 21719571 12364791 14747013 17210077
28004739 18362917 19820115 24267698 17994087 25315136 25348373 25830018 26108605 28648823 17550304 15632085 17510324 25474469 26999592 26823975 15804580 12177427 10731132 12537572 14610274 20075255 20566863 24508170 21347285 25244985 21282665 20798317 19060216 21719571 12364791 14747013 17210077
EMBL
AB167737
BAD23845.1
AK289335
BAM73862.1
AK289334
BAM73861.1
+ More
AK289333 BAM73860.1 DQ357068 ABC96070.1 KP764855 ALJ84053.1 AB649119 BAN66313.1 RSAL01000041 RVE50862.1 FJ844407 ACY92454.1 KX443478 ASO98053.1 ODYU01002692 SOQ40527.1 JTDY01000648 KOB76340.1 KK852775 KDR16797.1 GEZM01029690 JAV85990.1 LJIG01022600 KRT79716.1 KQ971343 EFA04669.1 NEVH01025175 PNF15482.1 KF044272 AGT57844.1 KX609520 ARN17927.1 CH964272 EDW85100.1 GAKP01014550 GAKP01014545 JAC44402.1 GBXI01007364 JAD06928.1 JRES01000630 KNC29778.1 KQ414632 KOC67115.1 ACPB03000489 NNAY01001009 OXU25500.1 GDHF01029511 JAI22803.1 CM000160 EDW97650.1 OUUW01000008 SPP83639.1 KZ288232 PBC31522.1 GFTR01006245 JAW10181.1 KQ978287 KYM95608.1 CM000070 EAL28793.2 CH477807 EAT36035.2 CH479185 EDW37990.1 GBBI01004133 JAC14579.1 GDUN01000232 JAN95687.1 KC515377 AGF44568.1 CH480815 EDW42545.1 GDHC01005469 JAQ13160.1 GEDC01020137 JAS17161.1 CH954181 EDV49458.1 AY947551 AAX85207.1 AY079170 AE014297 BT016061 CP012526 ALC47308.1 CM000364 EDX13308.1 AJWK01017455 GFXV01001329 MBW13134.1 DS235327 EEB14725.1 GDHC01010983 JAQ07646.1 KK107522 EZA49326.1 CH902623 EDV30217.1 JH431998 GAKP01014548 GAKP01014542 JAC44404.1 CH933806 EDW15638.1 CH916369 EDV92546.1 KQ982769 KYQ50946.1 JXJN01012594 CVRI01000054 CRL00351.1 KQ979479 KYN21367.1 GECZ01016961 GECZ01000749 JAS52808.1 JAS69020.1 ADTU01013744 ABLF02022089 CH940650 EDW66851.1 GL762454 EFZ21475.1 JH431820 GL444420 EFN60908.1 KQ981490 KYN41395.1 KQ976736 KYM76013.1 KK117489 KFM70608.1 EF546764 ABU42524.1 GL888103 EGI67350.1 AXCM01004618 AAAB01008846 EAA06503.5 GFTR01005892 JAW10534.1 GGMS01006436 MBY75639.1 KM216998 AIW79961.1 LBMM01006459 KMQ90601.1 AXCN02002153
AK289333 BAM73860.1 DQ357068 ABC96070.1 KP764855 ALJ84053.1 AB649119 BAN66313.1 RSAL01000041 RVE50862.1 FJ844407 ACY92454.1 KX443478 ASO98053.1 ODYU01002692 SOQ40527.1 JTDY01000648 KOB76340.1 KK852775 KDR16797.1 GEZM01029690 JAV85990.1 LJIG01022600 KRT79716.1 KQ971343 EFA04669.1 NEVH01025175 PNF15482.1 KF044272 AGT57844.1 KX609520 ARN17927.1 CH964272 EDW85100.1 GAKP01014550 GAKP01014545 JAC44402.1 GBXI01007364 JAD06928.1 JRES01000630 KNC29778.1 KQ414632 KOC67115.1 ACPB03000489 NNAY01001009 OXU25500.1 GDHF01029511 JAI22803.1 CM000160 EDW97650.1 OUUW01000008 SPP83639.1 KZ288232 PBC31522.1 GFTR01006245 JAW10181.1 KQ978287 KYM95608.1 CM000070 EAL28793.2 CH477807 EAT36035.2 CH479185 EDW37990.1 GBBI01004133 JAC14579.1 GDUN01000232 JAN95687.1 KC515377 AGF44568.1 CH480815 EDW42545.1 GDHC01005469 JAQ13160.1 GEDC01020137 JAS17161.1 CH954181 EDV49458.1 AY947551 AAX85207.1 AY079170 AE014297 BT016061 CP012526 ALC47308.1 CM000364 EDX13308.1 AJWK01017455 GFXV01001329 MBW13134.1 DS235327 EEB14725.1 GDHC01010983 JAQ07646.1 KK107522 EZA49326.1 CH902623 EDV30217.1 JH431998 GAKP01014548 GAKP01014542 JAC44404.1 CH933806 EDW15638.1 CH916369 EDV92546.1 KQ982769 KYQ50946.1 JXJN01012594 CVRI01000054 CRL00351.1 KQ979479 KYN21367.1 GECZ01016961 GECZ01000749 JAS52808.1 JAS69020.1 ADTU01013744 ABLF02022089 CH940650 EDW66851.1 GL762454 EFZ21475.1 JH431820 GL444420 EFN60908.1 KQ981490 KYN41395.1 KQ976736 KYM76013.1 KK117489 KFM70608.1 EF546764 ABU42524.1 GL888103 EGI67350.1 AXCM01004618 AAAB01008846 EAA06503.5 GFTR01005892 JAW10534.1 GGMS01006436 MBY75639.1 KM216998 AIW79961.1 LBMM01006459 KMQ90601.1 AXCN02002153
Proteomes
UP000283053
UP000037510
UP000027135
UP000007266
UP000235965
UP000078200
+ More
UP000092445 UP000007798 UP000095301 UP000005203 UP000037069 UP000053825 UP000015103 UP000215335 UP000095300 UP000002282 UP000268350 UP000242457 UP000078542 UP000001819 UP000008820 UP000008744 UP000001292 UP000008711 UP000000803 UP000092553 UP000000304 UP000002358 UP000092461 UP000009046 UP000053097 UP000192221 UP000007801 UP000091820 UP000009192 UP000001070 UP000075809 UP000092443 UP000092460 UP000075884 UP000183832 UP000078492 UP000005205 UP000007819 UP000076408 UP000008792 UP000000311 UP000078541 UP000078540 UP000054359 UP000075900 UP000007755 UP000075883 UP000075902 UP000007062 UP000036403 UP000075886 UP000075881 UP000075903
UP000092445 UP000007798 UP000095301 UP000005203 UP000037069 UP000053825 UP000015103 UP000215335 UP000095300 UP000002282 UP000268350 UP000242457 UP000078542 UP000001819 UP000008820 UP000008744 UP000001292 UP000008711 UP000000803 UP000092553 UP000000304 UP000002358 UP000092461 UP000009046 UP000053097 UP000192221 UP000007801 UP000091820 UP000009192 UP000001070 UP000075809 UP000092443 UP000092460 UP000075884 UP000183832 UP000078492 UP000005205 UP000007819 UP000076408 UP000008792 UP000000311 UP000078541 UP000078540 UP000054359 UP000075900 UP000007755 UP000075883 UP000075902 UP000007062 UP000036403 UP000075886 UP000075881 UP000075903
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
Q6I6M8
L0N6K4
L0N747
L0N7B2
Q2HZZ4
A0A0P0HKS9
+ More
S6B478 A0A3S2NLQ3 D1GEJ6 A0A248QEJ3 A0A2H1VI64 A0A0L7LLN4 A0A067RDD3 A0A1Y1MMR8 A0A0T6AXK2 D6WKZ9 A0A2J7PGM2 V5K5L7 A0A1W6L1B8 A0A1A9V720 A0A1A9ZL34 B4NK58 A0A1I8N6Z5 A0A034VQK8 A0A088APZ6 A0A0A1X8D5 A0A0L0CBJ1 A0A0L7R8B6 T1HQU7 A0A232F4V0 A0A0K8U898 A0A1I8Q149 B4PTF1 A0A3B0KBX2 A0A2A3EJT6 A0A224XQA4 A0A195C616 Q295B9 Q16NV1 B4GMB6 A0A023F0S2 A0A0P6IXI5 A0A075BFL1 B4HHW3 A0A146M3K4 A0A1B6CUW2 B3P4E3 Q563C5 Q9VGH1 A0A0M4EJV6 B4QTK3 K7IRZ9 A0A1B0CLM4 A0A2H8TGC2 E0VMW9 A0A146LLE3 A0A026W166 A0A1W4WC86 B3MTT6 A0A1A9X2R5 T1JAE9 A0A034VPJ7 B4K9Z0 B4JG07 A0A151WSX3 A0A1A9XW33 A0A1B0BDP7 A0A182N8F3 A0A1J1IJC8 A0A195E966 A0A1B6FRJ4 A0A158NF50 X1WI94 A0A182YH61 B4LXY5 E9ID25 T1J337 E2B079 A0A195FL49 A0A195AUU7 A0A087TZR9 A0A182RHK3 A9Q617 F4WEI7 A0A182MK14 A0A182U3F5 Q7QF34 A0A224XHL4 A0A2S2QD38 A0A0K0LB69 A0A0J7KK56 A0A182QLY6 A0A182KCS5 A0A182VHA7
S6B478 A0A3S2NLQ3 D1GEJ6 A0A248QEJ3 A0A2H1VI64 A0A0L7LLN4 A0A067RDD3 A0A1Y1MMR8 A0A0T6AXK2 D6WKZ9 A0A2J7PGM2 V5K5L7 A0A1W6L1B8 A0A1A9V720 A0A1A9ZL34 B4NK58 A0A1I8N6Z5 A0A034VQK8 A0A088APZ6 A0A0A1X8D5 A0A0L0CBJ1 A0A0L7R8B6 T1HQU7 A0A232F4V0 A0A0K8U898 A0A1I8Q149 B4PTF1 A0A3B0KBX2 A0A2A3EJT6 A0A224XQA4 A0A195C616 Q295B9 Q16NV1 B4GMB6 A0A023F0S2 A0A0P6IXI5 A0A075BFL1 B4HHW3 A0A146M3K4 A0A1B6CUW2 B3P4E3 Q563C5 Q9VGH1 A0A0M4EJV6 B4QTK3 K7IRZ9 A0A1B0CLM4 A0A2H8TGC2 E0VMW9 A0A146LLE3 A0A026W166 A0A1W4WC86 B3MTT6 A0A1A9X2R5 T1JAE9 A0A034VPJ7 B4K9Z0 B4JG07 A0A151WSX3 A0A1A9XW33 A0A1B0BDP7 A0A182N8F3 A0A1J1IJC8 A0A195E966 A0A1B6FRJ4 A0A158NF50 X1WI94 A0A182YH61 B4LXY5 E9ID25 T1J337 E2B079 A0A195FL49 A0A195AUU7 A0A087TZR9 A0A182RHK3 A9Q617 F4WEI7 A0A182MK14 A0A182U3F5 Q7QF34 A0A224XHL4 A0A2S2QD38 A0A0K0LB69 A0A0J7KK56 A0A182QLY6 A0A182KCS5 A0A182VHA7
PDB
3K9Y
E-value=3.28311e-46,
Score=467
Ontologies
GO
Topology
Subcellular location
Membrane
Mitochondrion membrane
Mitochondrion membrane
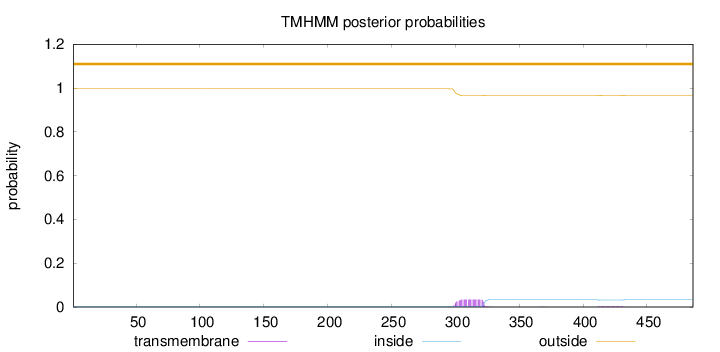
Length:
486
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.832939999999999
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00360
outside
1 - 486
Population Genetic Test Statistics
Pi
265.974022
Theta
190.589539
Tajima's D
1.618701
CLR
0.115263
CSRT
0.812609369531523
Interpretation
Uncertain
