Gene
KWMTBOMO04610 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005315
Annotation
PREDICTED:_26S_proteasome_non-ATPase_regulatory_subunit_2_isoform_X2_[Bombyx_mori]
Full name
26S proteasome non-ATPase regulatory subunit 2
Location in the cell
Cytoplasmic Reliability : 1.863
Sequence
CDS
ATGACTGTGAAAAATAAAACTGAAGAGCCAAAGAAGGAAAAAGCTGAGCCCGCAGTGGCTCCAAATGATGATCTTTCCGAGGAAGACAAACGCCTACAGGAGGAATTGAATATGCTAGTCGATAAGCTTTTGGGCAATGAAGTCGATTTATATTTTCCAGCATTACAAATGCTAAGTAATCTCATTCGGACGTCAACAACATCTATGACTTCTGTACCTAAACCTCTAAAATTCCTAAGGGAACATTATCCAGCATTGAAACAAGTATATGAAAAAATTACAGATGAAAAAACAAAAAAATTCTGTGCTGATGTTATCTCTGTCTTAGCGATGGGCGTAAGCGGTACTCTGGAAGTTGCAGAAAAGAGAGAGTGTTTGAAGTATTGTCTTCTGGGAACATTGTCAAATGTAGGTGATTGGGGACATGAATATGTTAGGCAATTGGAAGGTGAAATTGCTGAAGAATGGAATATAGAAAACATGGATTCACTTTTGCCACTAGTACGTGATGTTATCACATTTGACATGAAACACTCAGCTGAGATCCAGGCTTGTGATCTGTTAATGGAAATTGATAGACTTGATCTGCTGACCCAACATATGGACCAGAGCAACTACCCAAGAGTCTGTCTTTATCTTATTGGATGTGCAAGTTATGTTGTTGAGCCTGAGTCAACACAAATACTACAAGGTGTCTTAGATACATACCTTCGTTTTGGAGAGTACCCAAGAGCAATGCTTGTAGCTATGCAACTTCATGAGAAAGCTAAATGTGAAGAAGTGTTCAATGCCTGTAATGATCCTTTAATAAAAAAGCAACTGTGCTACTTACTTGCACGTCAATACATACCATTAGAAGTGGATGACGAAGATCTGCGCACAATTCTACTGAACGCTCATATCAATGATCATTTTCTAAGTCTGGCAAGAGAGCTTGATATTATGGAACCGAAAACTCCAGAAGAAGTTTATAAAACATGGTTAGAGTCGGCCGGATCAGCTCTTCGTCCATCTTTACTGACTGAACATCCTGTAGACTCTGCTAGACAGAACCTGTCAGCTACATTTGTGAACGCCTTTGTGAATGCCGGGTTTGGCCGAGACAAACTTGTCACAACCGATGATGGGAACAAATGGATGTACAAAAATAAGGATCATGGTATGCTCTCAGCGGCCGCGTCCCTTGGTATGATTCACTTGTGGGATGTAGACGGTGGTCTAACGCCTATCGACAAATACCTGTATACCGCCGACGAGCACATCAAGGCTGGTGCCCTACTGGCTTTAGGTCTCGTGAACTGCGGTGTCAGGAACGAGTGTGATCCAGCCTTAGCTCTGCTCTCCGATTATGTTCTGCATTCCAGTTCCAATCTCAGGATCGGAAGTGTTTTGGGTCTAGGTATTGCATATGCGGGAACACAACGAGAAGATGTGCTGTCCCACTTGCTTCCCGTGCTCAACGACACGACCGCCCCGGCCGAGATCTGCGCACTCGCCGCCATCTCCTGCGGGCTCATCGCCGTTGGATCGTGCAATGGCGATGTGTCTTGTGCAATCATTCAGCGATTGATTGACGATAACAAGGATTTGCACTCGTCTACTTACGCCAGGTTCCTGCATTTGGGACTCGGGCTGTGTTTCTTAGGCTGCAAGGAGCGCACGGAGGCGACGATGGCCGCGTTGGAAGTACTTCCGGAGCCGCAGCAGTCGCTCTGCCAGACCACGCTGTCCATGTGCGCGTACGCCGGCACCGGGGACGTGCTCGTGGTGCAACAGATGCTGCACATCTGCTCCAAGCACTACGAAACCGATAATGAACAATCGTCAACTGAAGACACAGCTTTTAAGAAACAAGACAAGAAGGACGCAAAAGAAACTACAGCAACTCCGAGCAGTTCAAAGGACGACAAATCCAAAGGTAAATCATCAAAAGACAGCAAAAACAAGGAAAAGGATAAGGAAAAGGAAGCCAACAAGGAGCTGTCCTCGGTGCAAGCCGTGGCGACCCTTGGCGTCGCGGTCATCGCATTGGCCGAGGAAACGGGCGCCGAAATGTGCACCCGCATCTTCGGACAACTGGGTAGGTACGGGGAGCCGGCAGTGCGGCGCGCCGTGCCGCTCGCCATCGCCCTGTGCTCCGTGTCCAACCCGCAGCTCTCCGTCATCGACGTGCTCAATAAATACTCCCACGATTCTGATAACGACGTCGCTTACAACGCCATATTCGCCATGGGACTCGTGGGCGCCGGCACTAACAACGCAAGACTGGCGACGATGCTGCGTGCGCTGGCGCTGTACCACGGCAAGTCTCCGGTGCACCTGTTCATGGTGCGGCTGGCGCAGGGGCTGTGTCACGCCGGCAAGGGCACGGTGACCCTGTGTCCCGCGCACGCGGACCGGCGCCTGCTCAACCAGCCCGCGCTCGCCGGCCTGCTCGTCGTGCTCACCGCCTTCCTCGACTGCAAGACCATAATCCTCGGCAAATCCCACTACTTGCTGTACGTATTGGCGACTGCGATGCAACCCCGCTGGCTGGTCACCCTCGACGAAAATCTTCAGCCCTTGAACGTCAGCGTGCGTGTGGGACAGGCTGTGGATGTTATCGGAAAAGCCGGTACACCGAAGACAATAGCCGGTTCCCACACTCACACGACGCCGGTGCTGCTGTCCTTCGGCGAGCGAGCAGAACTGGCGACTGACGAGTACATACCACTCTCGCCGATAATGGAGGGATTCGTGATTCTCAAGAAAAACGAGGATAGCGTCATGGCCTCTGTTCAATAA
Protein
MTVKNKTEEPKKEKAEPAVAPNDDLSEEDKRLQEELNMLVDKLLGNEVDLYFPALQMLSNLIRTSTTSMTSVPKPLKFLREHYPALKQVYEKITDEKTKKFCADVISVLAMGVSGTLEVAEKRECLKYCLLGTLSNVGDWGHEYVRQLEGEIAEEWNIENMDSLLPLVRDVITFDMKHSAEIQACDLLMEIDRLDLLTQHMDQSNYPRVCLYLIGCASYVVEPESTQILQGVLDTYLRFGEYPRAMLVAMQLHEKAKCEEVFNACNDPLIKKQLCYLLARQYIPLEVDDEDLRTILLNAHINDHFLSLARELDIMEPKTPEEVYKTWLESAGSALRPSLLTEHPVDSARQNLSATFVNAFVNAGFGRDKLVTTDDGNKWMYKNKDHGMLSAAASLGMIHLWDVDGGLTPIDKYLYTADEHIKAGALLALGLVNCGVRNECDPALALLSDYVLHSSSNLRIGSVLGLGIAYAGTQREDVLSHLLPVLNDTTAPAEICALAAISCGLIAVGSCNGDVSCAIIQRLIDDNKDLHSSTYARFLHLGLGLCFLGCKERTEATMAALEVLPEPQQSLCQTTLSMCAYAGTGDVLVVQQMLHICSKHYETDNEQSSTEDTAFKKQDKKDAKETTATPSSSKDDKSKGKSSKDSKNKEKDKEKEANKELSSVQAVATLGVAVIALAEETGAEMCTRIFGQLGRYGEPAVRRAVPLAIALCSVSNPQLSVIDVLNKYSHDSDNDVAYNAIFAMGLVGAGTNNARLATMLRALALYHGKSPVHLFMVRLAQGLCHAGKGTVTLCPAHADRRLLNQPALAGLLVVLTAFLDCKTIILGKSHYLLYVLATAMQPRWLVTLDENLQPLNVSVRVGQAVDVIGKAGTPKTIAGSHTHTTPVLLSFGERAELATDEYIPLSPIMEGFVILKKNEDSVMASVQ
Summary
Description
Component of the 26S proteasome, a multiprotein complex involved in the ATP-dependent degradation of ubiquitinated proteins. This complex plays a key role in the maintenance of protein homeostasis by removing misfolded or damaged proteins, which could impair cellular functions, and by removing proteins whose functions are no longer required. Therefore, the proteasome participates in numerous cellular processes, including cell cycle progression, apoptosis, or DNA damage repair.
Subunit
Component of the 19S proteasome regulatory particle complex. The 26S proteasome consists of a 20S core particle (CP) and two 19S regulatory subunits (RP).
Similarity
Belongs to the proteasome subunit S2 family.
Uniprot
H9J723
A0A2H1VIA1
A0A194PS70
A0A1E1WRU9
A0A194QSM7
S4NSS4
+ More
A0A1E1WMN7 T1HNA1 R4G4X6 A0A224X6X3 A0A0V0G3B6 A0A069DXQ6 A0A0A9XG19 A0A023F919 R4WTN1 A0A1S3D648 A0A1B6BXI5 A0A2H8TF17 A0A2S2P6A3 J9KAY2 V5HKA2 A0A1W4ZV99 A0A3B4DP07 A0A3B4BVP6 D6W687 T1JGZ6 A0A3Q3FT65 A0A2U9B5X4 A0A3B3SLU7 A0A1A7Z6M3 H2M068 A0A3P9IP80 A0A1A7WEI5 A0A3P9MJH0 A0A096MCN6 A0A1Y1LJJ1 A0A224YI86 F4WP73 E2BBF9 A0A060XQT1 C0H8V2 A0A195EV76 A0A1E1X2C0 A0A151WM61 Q5EB37 A0A1A8GYR1 W5MQF1 A0A3B3STW8 A0A3Q3M3J9 A0A060XNU2 A0A151ILX6 A0A131Z5I7 J3JXN2 A0A026WK09 A0A1S3T1T4 A0A3S2PEK4 A0A3Q1F3H4 F7CS21 A0A1A8HZY7 A0A3Q3G638 A0A195B112 A0A158NQW6 A0A3B3ST70 L7M501 A0A3B3TJA5 A0A131XMY5 A0A087Y778 A0A3B3DU11 A0A1A8C9D0 A0A1A8AKB1 A0A1A8RI97 A0A195E6A8 A0A195CMA8 A0A1E1XST9 A0A3Q3KNA7 A0A1A8MWD8 Q6PHK7 A0A023G054 R4GC62 K1PEY4 H9G5W8 K9J3W5 A0A2Y9DW10 A0A1S3A5Y3 A0A0L7QUA5 A0A2Y9F9K6 U3BFK1 A0A2R5LP41 E2RCP9 A0A2D0STZ9 A0A1S3GI88 W5U7P5 A0A0J7KMY8 A0A232F732 I6L5A0 F7EGU4 K7J2M6 A0A2L2XZG3 A0A0M9A0L6
A0A1E1WMN7 T1HNA1 R4G4X6 A0A224X6X3 A0A0V0G3B6 A0A069DXQ6 A0A0A9XG19 A0A023F919 R4WTN1 A0A1S3D648 A0A1B6BXI5 A0A2H8TF17 A0A2S2P6A3 J9KAY2 V5HKA2 A0A1W4ZV99 A0A3B4DP07 A0A3B4BVP6 D6W687 T1JGZ6 A0A3Q3FT65 A0A2U9B5X4 A0A3B3SLU7 A0A1A7Z6M3 H2M068 A0A3P9IP80 A0A1A7WEI5 A0A3P9MJH0 A0A096MCN6 A0A1Y1LJJ1 A0A224YI86 F4WP73 E2BBF9 A0A060XQT1 C0H8V2 A0A195EV76 A0A1E1X2C0 A0A151WM61 Q5EB37 A0A1A8GYR1 W5MQF1 A0A3B3STW8 A0A3Q3M3J9 A0A060XNU2 A0A151ILX6 A0A131Z5I7 J3JXN2 A0A026WK09 A0A1S3T1T4 A0A3S2PEK4 A0A3Q1F3H4 F7CS21 A0A1A8HZY7 A0A3Q3G638 A0A195B112 A0A158NQW6 A0A3B3ST70 L7M501 A0A3B3TJA5 A0A131XMY5 A0A087Y778 A0A3B3DU11 A0A1A8C9D0 A0A1A8AKB1 A0A1A8RI97 A0A195E6A8 A0A195CMA8 A0A1E1XST9 A0A3Q3KNA7 A0A1A8MWD8 Q6PHK7 A0A023G054 R4GC62 K1PEY4 H9G5W8 K9J3W5 A0A2Y9DW10 A0A1S3A5Y3 A0A0L7QUA5 A0A2Y9F9K6 U3BFK1 A0A2R5LP41 E2RCP9 A0A2D0STZ9 A0A1S3GI88 W5U7P5 A0A0J7KMY8 A0A232F732 I6L5A0 F7EGU4 K7J2M6 A0A2L2XZG3 A0A0M9A0L6
Pubmed
19121390
26354079
23622113
26334808
25401762
26823975
+ More
25474469 23691247 25765539 18362917 19820115 29240929 17554307 28004739 28797301 21719571 20798317 24755649 20433749 28503490 26830274 22516182 23537049 24508170 30249741 20431018 21347285 25576852 28049606 29451363 29209593 21881562 22992520 25243066 16341006 23127152 28648823 17495919 20075255 26561354
25474469 23691247 25765539 18362917 19820115 29240929 17554307 28004739 28797301 21719571 20798317 24755649 20433749 28503490 26830274 22516182 23537049 24508170 30249741 20431018 21347285 25576852 28049606 29451363 29209593 21881562 22992520 25243066 16341006 23127152 28648823 17495919 20075255 26561354
EMBL
BABH01013950
BABH01013951
ODYU01002692
SOQ40526.1
KQ459595
KPI95599.1
+ More
GDQN01001305 JAT89749.1 KQ461154 KPJ08528.1 GAIX01013937 JAA78623.1 GDQN01002867 JAT88187.1 ACPB03007651 GAHY01000990 JAA76520.1 GFTR01008250 JAW08176.1 GECL01003513 JAP02611.1 GBGD01000402 JAC88487.1 GBHO01023952 GBRD01009718 GDHC01011656 JAG19652.1 JAG56106.1 JAQ06973.1 GBBI01000751 JAC17961.1 AK418052 BAN21267.1 GEDC01031578 JAS05720.1 GFXV01000888 MBW12693.1 GGMR01012370 MBY24989.1 ABLF02033855 GANP01000328 JAB84140.1 KQ971306 EFA11079.1 JH432213 CP026245 AWO99245.1 HADX01015896 SBP38128.1 HADW01003002 SBP04402.1 AYCK01009338 AYCK01009339 GEZM01054149 JAV73819.1 GFPF01005529 MAA16675.1 GL888243 EGI64004.1 GL447043 EFN86966.1 FR905425 CDQ79230.1 BT058758 ACN10471.1 KQ981965 KYN31789.1 GFAC01005781 JAT93407.1 KQ982944 KYQ48954.1 BC090101 AAH90101.1 HAEB01018051 HAEC01008220 SBQ76358.1 AHAT01034287 FR905725 CDQ81233.1 KQ977085 KYN05887.1 GEDV01003176 JAP85381.1 APGK01047258 BT128002 KB741077 AEE62964.1 ENN74289.1 KK107182 QOIP01000003 EZA55991.1 RLU24575.1 CM012449 RVE64743.1 AAMC01042067 HAED01004959 HAEE01011452 SBQ90989.1 KQ976691 KYM77980.1 ADTU01023676 GACK01006870 JAA58164.1 GEFH01001670 JAP66911.1 HADZ01011460 HAEA01008187 SBP75401.1 HADY01017137 HAEJ01007360 SBP55622.1 HAEH01017201 HAEI01011234 SBS05746.1 KQ979608 KYN20412.1 KQ977574 KYN01825.1 GFAA01001365 JAU02070.1 HAEF01019655 HAEG01012876 SBR60814.1 BC056516 AAH56516.1 GBBL01000927 JAC26393.1 AAWZ02021625 JH818817 EKC22447.1 GABZ01000629 JAA52896.1 KQ414735 KOC62200.1 GAMT01000288 GAMR01000919 GAMP01003346 JAB11573.1 JAB33013.1 JAB49409.1 GGLE01006951 MBY11077.1 AAEX03017275 JT406941 AHH37790.1 LBMM01005329 KMQ91604.1 NNAY01000831 OXU26290.1 ABGA01216381 ABGA01216382 ABGA01216383 IAAA01002438 IAAA01002439 IAAA01002440 IAAA01002441 LAA01117.1 KQ435776 KOX74910.1
GDQN01001305 JAT89749.1 KQ461154 KPJ08528.1 GAIX01013937 JAA78623.1 GDQN01002867 JAT88187.1 ACPB03007651 GAHY01000990 JAA76520.1 GFTR01008250 JAW08176.1 GECL01003513 JAP02611.1 GBGD01000402 JAC88487.1 GBHO01023952 GBRD01009718 GDHC01011656 JAG19652.1 JAG56106.1 JAQ06973.1 GBBI01000751 JAC17961.1 AK418052 BAN21267.1 GEDC01031578 JAS05720.1 GFXV01000888 MBW12693.1 GGMR01012370 MBY24989.1 ABLF02033855 GANP01000328 JAB84140.1 KQ971306 EFA11079.1 JH432213 CP026245 AWO99245.1 HADX01015896 SBP38128.1 HADW01003002 SBP04402.1 AYCK01009338 AYCK01009339 GEZM01054149 JAV73819.1 GFPF01005529 MAA16675.1 GL888243 EGI64004.1 GL447043 EFN86966.1 FR905425 CDQ79230.1 BT058758 ACN10471.1 KQ981965 KYN31789.1 GFAC01005781 JAT93407.1 KQ982944 KYQ48954.1 BC090101 AAH90101.1 HAEB01018051 HAEC01008220 SBQ76358.1 AHAT01034287 FR905725 CDQ81233.1 KQ977085 KYN05887.1 GEDV01003176 JAP85381.1 APGK01047258 BT128002 KB741077 AEE62964.1 ENN74289.1 KK107182 QOIP01000003 EZA55991.1 RLU24575.1 CM012449 RVE64743.1 AAMC01042067 HAED01004959 HAEE01011452 SBQ90989.1 KQ976691 KYM77980.1 ADTU01023676 GACK01006870 JAA58164.1 GEFH01001670 JAP66911.1 HADZ01011460 HAEA01008187 SBP75401.1 HADY01017137 HAEJ01007360 SBP55622.1 HAEH01017201 HAEI01011234 SBS05746.1 KQ979608 KYN20412.1 KQ977574 KYN01825.1 GFAA01001365 JAU02070.1 HAEF01019655 HAEG01012876 SBR60814.1 BC056516 AAH56516.1 GBBL01000927 JAC26393.1 AAWZ02021625 JH818817 EKC22447.1 GABZ01000629 JAA52896.1 KQ414735 KOC62200.1 GAMT01000288 GAMR01000919 GAMP01003346 JAB11573.1 JAB33013.1 JAB49409.1 GGLE01006951 MBY11077.1 AAEX03017275 JT406941 AHH37790.1 LBMM01005329 KMQ91604.1 NNAY01000831 OXU26290.1 ABGA01216381 ABGA01216382 ABGA01216383 IAAA01002438 IAAA01002439 IAAA01002440 IAAA01002441 LAA01117.1 KQ435776 KOX74910.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000015103
UP000079169
UP000007819
+ More
UP000192224 UP000261440 UP000007266 UP000261660 UP000246464 UP000261540 UP000001038 UP000265200 UP000265180 UP000028760 UP000007755 UP000008237 UP000193380 UP000087266 UP000078541 UP000075809 UP000018468 UP000261640 UP000078542 UP000019118 UP000053097 UP000279307 UP000257200 UP000008143 UP000264800 UP000078540 UP000005205 UP000261500 UP000261560 UP000078492 UP000261600 UP000001646 UP000005408 UP000248480 UP000079721 UP000053825 UP000008225 UP000002254 UP000221080 UP000081671 UP000036403 UP000215335 UP000001595 UP000002280 UP000002358 UP000053105
UP000192224 UP000261440 UP000007266 UP000261660 UP000246464 UP000261540 UP000001038 UP000265200 UP000265180 UP000028760 UP000007755 UP000008237 UP000193380 UP000087266 UP000078541 UP000075809 UP000018468 UP000261640 UP000078542 UP000019118 UP000053097 UP000279307 UP000257200 UP000008143 UP000264800 UP000078540 UP000005205 UP000261500 UP000261560 UP000078492 UP000261600 UP000001646 UP000005408 UP000248480 UP000079721 UP000053825 UP000008225 UP000002254 UP000221080 UP000081671 UP000036403 UP000215335 UP000001595 UP000002280 UP000002358 UP000053105
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J723
A0A2H1VIA1
A0A194PS70
A0A1E1WRU9
A0A194QSM7
S4NSS4
+ More
A0A1E1WMN7 T1HNA1 R4G4X6 A0A224X6X3 A0A0V0G3B6 A0A069DXQ6 A0A0A9XG19 A0A023F919 R4WTN1 A0A1S3D648 A0A1B6BXI5 A0A2H8TF17 A0A2S2P6A3 J9KAY2 V5HKA2 A0A1W4ZV99 A0A3B4DP07 A0A3B4BVP6 D6W687 T1JGZ6 A0A3Q3FT65 A0A2U9B5X4 A0A3B3SLU7 A0A1A7Z6M3 H2M068 A0A3P9IP80 A0A1A7WEI5 A0A3P9MJH0 A0A096MCN6 A0A1Y1LJJ1 A0A224YI86 F4WP73 E2BBF9 A0A060XQT1 C0H8V2 A0A195EV76 A0A1E1X2C0 A0A151WM61 Q5EB37 A0A1A8GYR1 W5MQF1 A0A3B3STW8 A0A3Q3M3J9 A0A060XNU2 A0A151ILX6 A0A131Z5I7 J3JXN2 A0A026WK09 A0A1S3T1T4 A0A3S2PEK4 A0A3Q1F3H4 F7CS21 A0A1A8HZY7 A0A3Q3G638 A0A195B112 A0A158NQW6 A0A3B3ST70 L7M501 A0A3B3TJA5 A0A131XMY5 A0A087Y778 A0A3B3DU11 A0A1A8C9D0 A0A1A8AKB1 A0A1A8RI97 A0A195E6A8 A0A195CMA8 A0A1E1XST9 A0A3Q3KNA7 A0A1A8MWD8 Q6PHK7 A0A023G054 R4GC62 K1PEY4 H9G5W8 K9J3W5 A0A2Y9DW10 A0A1S3A5Y3 A0A0L7QUA5 A0A2Y9F9K6 U3BFK1 A0A2R5LP41 E2RCP9 A0A2D0STZ9 A0A1S3GI88 W5U7P5 A0A0J7KMY8 A0A232F732 I6L5A0 F7EGU4 K7J2M6 A0A2L2XZG3 A0A0M9A0L6
A0A1E1WMN7 T1HNA1 R4G4X6 A0A224X6X3 A0A0V0G3B6 A0A069DXQ6 A0A0A9XG19 A0A023F919 R4WTN1 A0A1S3D648 A0A1B6BXI5 A0A2H8TF17 A0A2S2P6A3 J9KAY2 V5HKA2 A0A1W4ZV99 A0A3B4DP07 A0A3B4BVP6 D6W687 T1JGZ6 A0A3Q3FT65 A0A2U9B5X4 A0A3B3SLU7 A0A1A7Z6M3 H2M068 A0A3P9IP80 A0A1A7WEI5 A0A3P9MJH0 A0A096MCN6 A0A1Y1LJJ1 A0A224YI86 F4WP73 E2BBF9 A0A060XQT1 C0H8V2 A0A195EV76 A0A1E1X2C0 A0A151WM61 Q5EB37 A0A1A8GYR1 W5MQF1 A0A3B3STW8 A0A3Q3M3J9 A0A060XNU2 A0A151ILX6 A0A131Z5I7 J3JXN2 A0A026WK09 A0A1S3T1T4 A0A3S2PEK4 A0A3Q1F3H4 F7CS21 A0A1A8HZY7 A0A3Q3G638 A0A195B112 A0A158NQW6 A0A3B3ST70 L7M501 A0A3B3TJA5 A0A131XMY5 A0A087Y778 A0A3B3DU11 A0A1A8C9D0 A0A1A8AKB1 A0A1A8RI97 A0A195E6A8 A0A195CMA8 A0A1E1XST9 A0A3Q3KNA7 A0A1A8MWD8 Q6PHK7 A0A023G054 R4GC62 K1PEY4 H9G5W8 K9J3W5 A0A2Y9DW10 A0A1S3A5Y3 A0A0L7QUA5 A0A2Y9F9K6 U3BFK1 A0A2R5LP41 E2RCP9 A0A2D0STZ9 A0A1S3GI88 W5U7P5 A0A0J7KMY8 A0A232F732 I6L5A0 F7EGU4 K7J2M6 A0A2L2XZG3 A0A0M9A0L6
PDB
6MSD
E-value=0,
Score=2463
Ontologies
GO
PANTHER
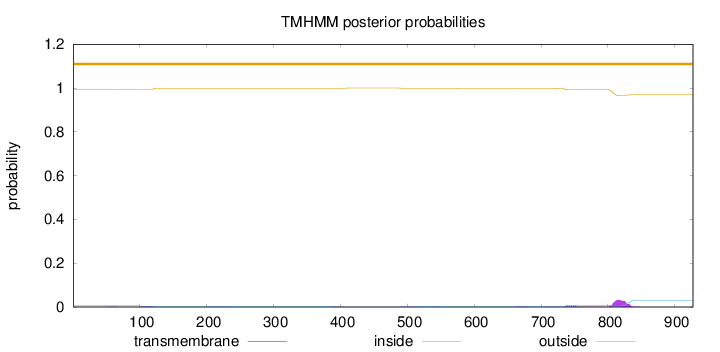
Topology
Length:
927
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.04302
Exp number, first 60 AAs:
0.00218
Total prob of N-in:
0.00612
outside
1 - 927
Population Genetic Test Statistics
Pi
249.41129
Theta
151.315049
Tajima's D
1.582094
CLR
0.008827
CSRT
0.810559472026399
Interpretation
Uncertain
