Gene
KWMTBOMO04607
Pre Gene Modal
BGIBMGA005316
Annotation
PREDICTED:_uncharacterized_protein_LOC101746213_[Bombyx_mori]
Full name
Deoxyhypusine hydroxylase
Alternative Name
Deoxyhypusine dioxygenase
Deoxyhypusine monooxygenase
Deoxyhypusine monooxygenase
Location in the cell
Nuclear Reliability : 1.978
Sequence
CDS
ATGGTTCGATTGTCAATAATTCTAAATAGAATTCCTGTTATAAAATTTCGTAAAGGTGGGGCTAGTCATGTCGGTACAAGCTCTCCCGGAGCTTCAGGACCCACTGCTGCTGCTACTGCTGGAACCCCTGTGCAAAGCCAGCCTGCTGCAGCAATGAGTACTGGTGCTATACCAGACATAGACCTACCAGCACGTTACAAGAGACAACCACTCTCAGAAGAAGAGATTGCTTATATCAATGGAGGAGGAATTGTTTAG
Protein
MVRLSIILNRIPVIKFRKGGASHVGTSSPGASGPTAAATAGTPVQSQPAAAMSTGAIPDIDLPARYKRQPLSEEEIAYINGGGIV
Summary
Description
Catalyzes the hydroxylation of the N(6)-(4-aminobutyl)-L-lysine intermediate to form hypusine, an essential post-translational modification only found in mature eIF-5A factor.
Catalytic Activity
[eIF5A protein]-deoxyhypusine + AH2 + O2 = [eIF5A protein]-hypusine + A + H2O
Cofactor
Fe(2+)
Similarity
Belongs to the deoxyhypusine hydroxylase family.
Uniprot
H9J724
A0A2A4IZB0
A0A2H1VWF7
A0A3S2M416
S4P3T7
A0A1B0F963
+ More
A0A1A9WHX0 A0A1B0BQN6 A0A0Q9XLT7 A0A0R1DT11 A0A1W4UZI3 A0A212EYR0 A0A1Q3FGM5 A0A1Q3FGG8 A0A1Q3FFW9 A0A0Q9W3D2 B4QHH1 A0A0J9U3F3 A0A0Q5VM39 Q2VKK0 Q2VKJ4 G2J5Z0 A0A1W4VC73 A0A1L8EEX2 U5ESB8 A0A158NDK1 A0A0L0C819 Q2VKI7 A0A1I8NCZ5 A0A0J9RE52 B7YZH7 A0A0R1DSN2 A0A2A3EH50 B0W2R3 T1E3F6 G2J5Y4 A0A023EE72 A0A0P8Y9Z4 A0A088A2L5 A0A1B0D8T7 A0A1B0CB85 A0A1L8EHI1 B4JW97 A0A1S4EWV3 A0A0Q5VN31 Q17N73 A0A0Q9WS23 T1E7Q3 W5JVG9 A0A2M4AKG8 A0A1Q3FHW0 A0A1J1IGM7 A0A1Q3FH20 A0A139WG59 A0A195BP30 A0A2M3ZKP3 A0A1I8NCZ0 A0A2M4AL05 A0A2M4ANR7 A0A2M4C5A7 A0A182R4P2 A0A182Y9D2 A0A2M3ZE99 A0A023ED80 K7IS90 A0A182QCI9 A0A1I8PP15 A0A0K8TRL0 A0A182N3Y9 A0A151XEE3 A0A164LPY0 A0A195CMY3 A0A195D9Y0 A0A182WDW2 A0A3L8E0A3 A0A226EY09 A0A1L8DSB1 A0A182JLK1 A0A1L8DR58 A0A0Q9X2Q8 A0A154PGU7 E2AQQ3 A0A0N0BG38 A0A0L7RB26 A0A084WIS0 B5E017 B4GAM3 F4W531 A0A182FUQ1 A0A026W6S7 A0A182HHG6 A0A182P4Y8 A0A1L8E849 A0A210QI14 A0A1D2MYH0
A0A1A9WHX0 A0A1B0BQN6 A0A0Q9XLT7 A0A0R1DT11 A0A1W4UZI3 A0A212EYR0 A0A1Q3FGM5 A0A1Q3FGG8 A0A1Q3FFW9 A0A0Q9W3D2 B4QHH1 A0A0J9U3F3 A0A0Q5VM39 Q2VKK0 Q2VKJ4 G2J5Z0 A0A1W4VC73 A0A1L8EEX2 U5ESB8 A0A158NDK1 A0A0L0C819 Q2VKI7 A0A1I8NCZ5 A0A0J9RE52 B7YZH7 A0A0R1DSN2 A0A2A3EH50 B0W2R3 T1E3F6 G2J5Y4 A0A023EE72 A0A0P8Y9Z4 A0A088A2L5 A0A1B0D8T7 A0A1B0CB85 A0A1L8EHI1 B4JW97 A0A1S4EWV3 A0A0Q5VN31 Q17N73 A0A0Q9WS23 T1E7Q3 W5JVG9 A0A2M4AKG8 A0A1Q3FHW0 A0A1J1IGM7 A0A1Q3FH20 A0A139WG59 A0A195BP30 A0A2M3ZKP3 A0A1I8NCZ0 A0A2M4AL05 A0A2M4ANR7 A0A2M4C5A7 A0A182R4P2 A0A182Y9D2 A0A2M3ZE99 A0A023ED80 K7IS90 A0A182QCI9 A0A1I8PP15 A0A0K8TRL0 A0A182N3Y9 A0A151XEE3 A0A164LPY0 A0A195CMY3 A0A195D9Y0 A0A182WDW2 A0A3L8E0A3 A0A226EY09 A0A1L8DSB1 A0A182JLK1 A0A1L8DR58 A0A0Q9X2Q8 A0A154PGU7 E2AQQ3 A0A0N0BG38 A0A0L7RB26 A0A084WIS0 B5E017 B4GAM3 F4W531 A0A182FUQ1 A0A026W6S7 A0A182HHG6 A0A182P4Y8 A0A1L8E849 A0A210QI14 A0A1D2MYH0
EC Number
1.14.99.29
Pubmed
19121390
23622113
17994087
17550304
22118469
22936249
+ More
16085702 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 26108605 25315136 24330624 24945155 26483478 17510324 20920257 23761445 18362917 19820115 25244985 20075255 26369729 30249741 20798317 24438588 15632085 21719571 24508170 28812685 27289101
16085702 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 26108605 25315136 24330624 24945155 26483478 17510324 20920257 23761445 18362917 19820115 25244985 20075255 26369729 30249741 20798317 24438588 15632085 21719571 24508170 28812685 27289101
EMBL
BABH01013949
NWSH01004892
PCG64604.1
ODYU01004863
SOQ45177.1
RSAL01000041
+ More
RVE50857.1 GAIX01006299 JAA86261.1 CCAG010022078 JXJN01018704 CH933808 KRG04739.1 CM000158 KRK00229.1 AGBW02011442 OWR46638.1 GFDL01008339 JAV26706.1 GFDL01008399 JAV26646.1 GFDL01008597 JAV26448.1 CH940648 KRF79378.1 CM000362 EDX07316.1 CM002911 KMY94200.1 CH954179 KQS62467.1 DQ079170 DQ079171 DQ079172 DQ079173 DQ079174 DQ079175 DQ079176 AAZ42654.1 AAZ42655.1 AAZ42656.1 AAZ42657.1 AAZ42658.1 AAZ42659.1 AAZ42660.1 DQ079177 DQ079178 DQ079179 DQ079180 DQ079181 DQ079182 DQ079183 AAZ42661.1 AAZ42662.1 AAZ42663.1 AAZ42664.1 AAZ42665.1 AAZ42666.1 AAZ42667.1 KRG04740.1 AE013599 BT128847 KX531797 ADV37203.1 AEM98445.1 ANY27607.1 GFDG01001558 JAV17241.1 GANO01003314 JAB56557.1 ADTU01012643 JRES01000883 KNC27564.1 DQ079184 AAZ42668.1 KMY94201.1 BT125795 ACL83130.1 ADR66769.1 KRK00230.1 KZ288252 PBC31038.1 DS231828 EDS29816.1 GALA01000400 JAA94452.1 BT128841 AEM92672.1 JXUM01147457 GAPW01006669 KQ570372 JAC06929.1 KXJ68376.1 CH902619 KPU75937.1 AJVK01027725 AJWK01004883 GFDG01000723 JAV18076.1 CH916375 EDV98235.1 KQS62468.1 CH477201 EAT48181.1 CH964154 KRF99087.1 GAMD01003453 JAA98137.1 ADMH02000368 ETN66809.1 GGFK01007965 MBW41286.1 GFDL01007889 JAV27156.1 CVRI01000047 CRK98198.1 GFDL01008242 JAV26803.1 KQ971351 KYB26787.1 KQ976424 KYM88270.1 GGFM01008386 MBW29137.1 GGFK01008155 MBW41476.1 GGFK01009115 MBW42436.1 GGFJ01011190 MBW60331.1 GGFM01006057 MBW26808.1 GAPW01006792 JAC06806.1 AXCN02001095 GDAI01000599 JAI17004.1 KQ982254 KYQ58742.1 LRGB01003123 KZS04327.1 KQ977600 KYN01449.1 KQ981082 KYN09667.1 QOIP01000002 RLU25913.1 LNIX01000001 OXA61721.1 GFDF01004755 JAV09329.1 GFDF01005132 JAV08952.1 KRF99088.1 KQ434899 KZC11027.1 GL441804 EFN64242.1 KQ435793 KOX74242.1 KQ414617 KOC68122.1 ATLV01023945 KE525347 KFB50114.1 CM000071 EDY68901.1 CH479181 EDW31975.1 GL887596 EGI70727.1 KK107399 EZA51331.1 APCN01002408 GFDG01003882 JAV14917.1 NEDP02003547 OWF48428.1 LJIJ01000418 ODM97705.1
RVE50857.1 GAIX01006299 JAA86261.1 CCAG010022078 JXJN01018704 CH933808 KRG04739.1 CM000158 KRK00229.1 AGBW02011442 OWR46638.1 GFDL01008339 JAV26706.1 GFDL01008399 JAV26646.1 GFDL01008597 JAV26448.1 CH940648 KRF79378.1 CM000362 EDX07316.1 CM002911 KMY94200.1 CH954179 KQS62467.1 DQ079170 DQ079171 DQ079172 DQ079173 DQ079174 DQ079175 DQ079176 AAZ42654.1 AAZ42655.1 AAZ42656.1 AAZ42657.1 AAZ42658.1 AAZ42659.1 AAZ42660.1 DQ079177 DQ079178 DQ079179 DQ079180 DQ079181 DQ079182 DQ079183 AAZ42661.1 AAZ42662.1 AAZ42663.1 AAZ42664.1 AAZ42665.1 AAZ42666.1 AAZ42667.1 KRG04740.1 AE013599 BT128847 KX531797 ADV37203.1 AEM98445.1 ANY27607.1 GFDG01001558 JAV17241.1 GANO01003314 JAB56557.1 ADTU01012643 JRES01000883 KNC27564.1 DQ079184 AAZ42668.1 KMY94201.1 BT125795 ACL83130.1 ADR66769.1 KRK00230.1 KZ288252 PBC31038.1 DS231828 EDS29816.1 GALA01000400 JAA94452.1 BT128841 AEM92672.1 JXUM01147457 GAPW01006669 KQ570372 JAC06929.1 KXJ68376.1 CH902619 KPU75937.1 AJVK01027725 AJWK01004883 GFDG01000723 JAV18076.1 CH916375 EDV98235.1 KQS62468.1 CH477201 EAT48181.1 CH964154 KRF99087.1 GAMD01003453 JAA98137.1 ADMH02000368 ETN66809.1 GGFK01007965 MBW41286.1 GFDL01007889 JAV27156.1 CVRI01000047 CRK98198.1 GFDL01008242 JAV26803.1 KQ971351 KYB26787.1 KQ976424 KYM88270.1 GGFM01008386 MBW29137.1 GGFK01008155 MBW41476.1 GGFK01009115 MBW42436.1 GGFJ01011190 MBW60331.1 GGFM01006057 MBW26808.1 GAPW01006792 JAC06806.1 AXCN02001095 GDAI01000599 JAI17004.1 KQ982254 KYQ58742.1 LRGB01003123 KZS04327.1 KQ977600 KYN01449.1 KQ981082 KYN09667.1 QOIP01000002 RLU25913.1 LNIX01000001 OXA61721.1 GFDF01004755 JAV09329.1 GFDF01005132 JAV08952.1 KRF99088.1 KQ434899 KZC11027.1 GL441804 EFN64242.1 KQ435793 KOX74242.1 KQ414617 KOC68122.1 ATLV01023945 KE525347 KFB50114.1 CM000071 EDY68901.1 CH479181 EDW31975.1 GL887596 EGI70727.1 KK107399 EZA51331.1 APCN01002408 GFDG01003882 JAV14917.1 NEDP02003547 OWF48428.1 LJIJ01000418 ODM97705.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000092444
UP000091820
UP000092460
+ More
UP000009192 UP000002282 UP000192221 UP000007151 UP000008792 UP000000304 UP000008711 UP000000803 UP000005205 UP000037069 UP000095301 UP000242457 UP000002320 UP000069940 UP000249989 UP000007801 UP000005203 UP000092462 UP000092461 UP000001070 UP000008820 UP000007798 UP000000673 UP000183832 UP000007266 UP000078540 UP000075900 UP000076408 UP000002358 UP000075886 UP000095300 UP000075884 UP000075809 UP000076858 UP000078542 UP000078492 UP000075920 UP000279307 UP000198287 UP000075880 UP000076502 UP000000311 UP000053105 UP000053825 UP000030765 UP000001819 UP000008744 UP000007755 UP000069272 UP000053097 UP000075840 UP000075885 UP000242188 UP000094527
UP000009192 UP000002282 UP000192221 UP000007151 UP000008792 UP000000304 UP000008711 UP000000803 UP000005205 UP000037069 UP000095301 UP000242457 UP000002320 UP000069940 UP000249989 UP000007801 UP000005203 UP000092462 UP000092461 UP000001070 UP000008820 UP000007798 UP000000673 UP000183832 UP000007266 UP000078540 UP000075900 UP000076408 UP000002358 UP000075886 UP000095300 UP000075884 UP000075809 UP000076858 UP000078542 UP000078492 UP000075920 UP000279307 UP000198287 UP000075880 UP000076502 UP000000311 UP000053105 UP000053825 UP000030765 UP000001819 UP000008744 UP000007755 UP000069272 UP000053097 UP000075840 UP000075885 UP000242188 UP000094527
Pfam
PF10937 S36_mt
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
H9J724
A0A2A4IZB0
A0A2H1VWF7
A0A3S2M416
S4P3T7
A0A1B0F963
+ More
A0A1A9WHX0 A0A1B0BQN6 A0A0Q9XLT7 A0A0R1DT11 A0A1W4UZI3 A0A212EYR0 A0A1Q3FGM5 A0A1Q3FGG8 A0A1Q3FFW9 A0A0Q9W3D2 B4QHH1 A0A0J9U3F3 A0A0Q5VM39 Q2VKK0 Q2VKJ4 G2J5Z0 A0A1W4VC73 A0A1L8EEX2 U5ESB8 A0A158NDK1 A0A0L0C819 Q2VKI7 A0A1I8NCZ5 A0A0J9RE52 B7YZH7 A0A0R1DSN2 A0A2A3EH50 B0W2R3 T1E3F6 G2J5Y4 A0A023EE72 A0A0P8Y9Z4 A0A088A2L5 A0A1B0D8T7 A0A1B0CB85 A0A1L8EHI1 B4JW97 A0A1S4EWV3 A0A0Q5VN31 Q17N73 A0A0Q9WS23 T1E7Q3 W5JVG9 A0A2M4AKG8 A0A1Q3FHW0 A0A1J1IGM7 A0A1Q3FH20 A0A139WG59 A0A195BP30 A0A2M3ZKP3 A0A1I8NCZ0 A0A2M4AL05 A0A2M4ANR7 A0A2M4C5A7 A0A182R4P2 A0A182Y9D2 A0A2M3ZE99 A0A023ED80 K7IS90 A0A182QCI9 A0A1I8PP15 A0A0K8TRL0 A0A182N3Y9 A0A151XEE3 A0A164LPY0 A0A195CMY3 A0A195D9Y0 A0A182WDW2 A0A3L8E0A3 A0A226EY09 A0A1L8DSB1 A0A182JLK1 A0A1L8DR58 A0A0Q9X2Q8 A0A154PGU7 E2AQQ3 A0A0N0BG38 A0A0L7RB26 A0A084WIS0 B5E017 B4GAM3 F4W531 A0A182FUQ1 A0A026W6S7 A0A182HHG6 A0A182P4Y8 A0A1L8E849 A0A210QI14 A0A1D2MYH0
A0A1A9WHX0 A0A1B0BQN6 A0A0Q9XLT7 A0A0R1DT11 A0A1W4UZI3 A0A212EYR0 A0A1Q3FGM5 A0A1Q3FGG8 A0A1Q3FFW9 A0A0Q9W3D2 B4QHH1 A0A0J9U3F3 A0A0Q5VM39 Q2VKK0 Q2VKJ4 G2J5Z0 A0A1W4VC73 A0A1L8EEX2 U5ESB8 A0A158NDK1 A0A0L0C819 Q2VKI7 A0A1I8NCZ5 A0A0J9RE52 B7YZH7 A0A0R1DSN2 A0A2A3EH50 B0W2R3 T1E3F6 G2J5Y4 A0A023EE72 A0A0P8Y9Z4 A0A088A2L5 A0A1B0D8T7 A0A1B0CB85 A0A1L8EHI1 B4JW97 A0A1S4EWV3 A0A0Q5VN31 Q17N73 A0A0Q9WS23 T1E7Q3 W5JVG9 A0A2M4AKG8 A0A1Q3FHW0 A0A1J1IGM7 A0A1Q3FH20 A0A139WG59 A0A195BP30 A0A2M3ZKP3 A0A1I8NCZ0 A0A2M4AL05 A0A2M4ANR7 A0A2M4C5A7 A0A182R4P2 A0A182Y9D2 A0A2M3ZE99 A0A023ED80 K7IS90 A0A182QCI9 A0A1I8PP15 A0A0K8TRL0 A0A182N3Y9 A0A151XEE3 A0A164LPY0 A0A195CMY3 A0A195D9Y0 A0A182WDW2 A0A3L8E0A3 A0A226EY09 A0A1L8DSB1 A0A182JLK1 A0A1L8DR58 A0A0Q9X2Q8 A0A154PGU7 E2AQQ3 A0A0N0BG38 A0A0L7RB26 A0A084WIS0 B5E017 B4GAM3 F4W531 A0A182FUQ1 A0A026W6S7 A0A182HHG6 A0A182P4Y8 A0A1L8E849 A0A210QI14 A0A1D2MYH0
Ontologies
PANTHER
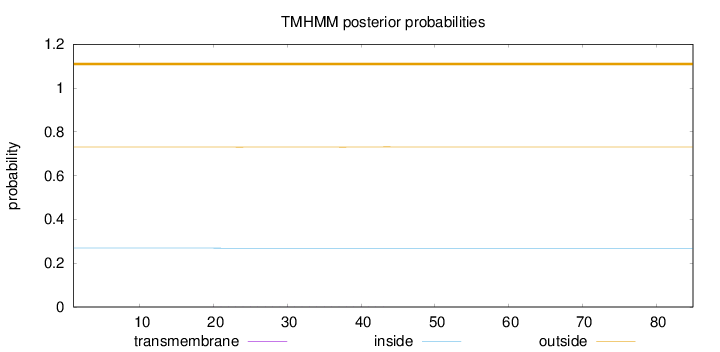
Topology
Length:
85
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01841
Exp number, first 60 AAs:
0.01841
Total prob of N-in:
0.26960
outside
1 - 85
Population Genetic Test Statistics
Pi
225.278952
Theta
214.379598
Tajima's D
-0.227611
CLR
0.976851
CSRT
0.312384380780961
Interpretation
Uncertain
