Pre Gene Modal
BGIBMGA005317
Annotation
PREDICTED:_THO_complex_subunit_5_homolog_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.903
Sequence
CDS
ATGGGAAAAGAAGAAGTAACGGCTGTTAAGAAACGGCGCAAATTAAATACAAGTACATCAAATGACGGAACAAAGCCAGTTCCCGTTGATGTATACAAGCGTGTTGTTGAATTTGAAGAAGCTGAAGCTCATCTGCGTCCAGCTGATAAGGATGCTGCCTTTTTCAATAAAACATGCCAAGACATAAGGCAACTTTTTAAAGAGATTGCAGAGTTAAAAACTAAAGAAACATTGGAGGCTAAAGAAAAGATAAATGCAAAAAGAGTGGAAGCTTCATTACTGCTGGTTGCGTTAAAGAAATTAAATCGCCTGGAGAAAGTACGAACTAGAGCTGGTCGTGATGCGCTCCATAAAGAGAAACAGAGAGTTGATTCCACACATTTACTGTTACAAAATTTACTTTACGAAGCAGATCACTTAAACAAAGAAGTAACTAAATGTTTACAATTCAAATCTAAAGATGAAGAAATAGATCTTGTGCCTCTTGAAGAATTCTATAAAAATGCACCTCCTGATATTATTAGATCAGAGGTCACAAAGGCTGATGAACATCAATTACAATTAGCCCGACTGGAGTGGGAGTTACGTCAACGTCGTGAATTATCTGAAGCCTGTAATGAACTCATATCTTCCAAGGAACGAGTAGCAGCTGCCATTGCTGCTGCACGTTCTAGACTTGATGCATTGGCTCCACATCTGAAAGATGTCTTGAAATCCACTAAACCTTTGCAAGAATGTCTTGCTCTACGCTTGGATGAAAAACGTGATGAAGCCAGGGCCGCAAGCTTGCTTCCAGCATCACTATTCCTACTATTTAAAAATGCAAACGCATATTCTGATATTTTAGGAAGCAAAGTTATTAGTGTTGGTGTTTGCGGTGATGAAGATGAAGCTCGCCGGCATGATCAAAACAATGTTGAAAATGATATTATTTCATCAAATGAGTCAGATTCAGATCAAGAAAATAATGATGAAGATCAACAAACCAAAAAGAAACGACATCACAGAACTAAAAACGTATCTAAAGAAGACAAAGCAGAAGCTAAAATAAAACGAGTATTCAAGAAACATCCTTTAAATGTACAAGTTTCAGTTAAAGCAGAAGATGGTACTGCATTGAATCTCATATTATCATACCTCATAAACTTGAAAATTATAGTTGTGAAGTTTACTGTATCGCTACCCAAACCTGTCACTGGTGTATCTGCTGCTGATGTTCTAAATGGACATTCTATTTTAAATGAACTGTACCCAGGGGATACGGGAGATGATTCACCCCACCCTGCAACTTCTTATTTACTAAAAAATGCAGGTATATCGGAAAGCTTTTCTTATTTCATACCAGAAATTGGACGCCCATACGTTTGGGCACAAAGAATGTGTGGAATTGATTTTATGACTTCAGAATTGAATGAGCAATCTGTTAGAAGAGCTGTTGAACCGTGTCCTAATCTCAGTGTCGCCACTGTTGAAAATTTTATCATGACATTGAAAAAGAGACTAAAATGTAGAGTAAAACTTATGCATGAGCTACAAGATCTAGAATCAGGAAAGATTTCACCACCAAAAGGAAGCTTGCCTCCTTCTATGCGTCTAGCAGGTTCTTTGACTCAGTGGCAGTCTGTTGGTTGGCCTGAATACAGTCAGGCTCCATCAACAGCTTTTCTTGTAACTGAAGGTTTAGTATCTGCAAATGATTTACTTTATAGAGCGATAATTACCAGACAATCAGCTAAGTTGGTGGCTTTAGTTGCCCTCAGAAGCGATTATCCTAAGAGCGTGCCAATATTTTCTCTAACTTTACACTGGAATGGCACTCATCACGCAGGTATTAATGATGATATCCGGGATATTGAAAGAGTCGTAAATACTGAGATAGGTTTAGAGGAACGCAAGCACTGCACCCTTTCTACTCAAATAACAAAATTATTAGCCTGTCTTGATATTCTATTGGAAACAAGCGATTCCCCAGAATTTCCACCAGACAAAGTTATACTTCATCCAGTTAGAGGACGTAACAGGAGCAAGCCATATAAATTTGTAAACCAAGGTGTTGGTGTCTTTGTACATCACTGA
Protein
MGKEEVTAVKKRRKLNTSTSNDGTKPVPVDVYKRVVEFEEAEAHLRPADKDAAFFNKTCQDIRQLFKEIAELKTKETLEAKEKINAKRVEASLLLVALKKLNRLEKVRTRAGRDALHKEKQRVDSTHLLLQNLLYEADHLNKEVTKCLQFKSKDEEIDLVPLEEFYKNAPPDIIRSEVTKADEHQLQLARLEWELRQRRELSEACNELISSKERVAAAIAAARSRLDALAPHLKDVLKSTKPLQECLALRLDEKRDEARAASLLPASLFLLFKNANAYSDILGSKVISVGVCGDEDEARRHDQNNVENDIISSNESDSDQENNDEDQQTKKKRHHRTKNVSKEDKAEAKIKRVFKKHPLNVQVSVKAEDGTALNLILSYLINLKIIVVKFTVSLPKPVTGVSAADVLNGHSILNELYPGDTGDDSPHPATSYLLKNAGISESFSYFIPEIGRPYVWAQRMCGIDFMTSELNEQSVRRAVEPCPNLSVATVENFIMTLKKRLKCRVKLMHELQDLESGKISPPKGSLPPSMRLAGSLTQWQSVGWPEYSQAPSTAFLVTEGLVSANDLLYRAIITRQSAKLVALVALRSDYPKSVPIFSLTLHWNGTHHAGINDDIRDIERVVNTEIGLEERKHCTLSTQITKLLACLDILLETSDSPEFPPDKVILHPVRGRNRSKPYKFVNQGVGVFVHH
Summary
Uniprot
H9J725
A0A2H1VWJ8
A0A3S2PGM4
A0A194PWT4
A0A194QT81
A0A212EYR7
+ More
A0A067QMF0 A0A2J7RRE6 F4WXM1 A0A151WSW5 A0A195FQK8 A0A026W386 A0A151JR84 A0A195BXH5 A0A195CRL6 A0A158NUX2 A0A336KZH7 A0A1B6MH52 A0A182JCL4 A0A336LT34 A0A1B6ENV1 D6WXN9 E2BKD1 E9ID99 A0A182G622 A0A1B0DAS6 A0A088AJP7 U5EKA3 A0A084VZT0 A0A2A3E2Y0 A0A1L8DCK4 A0A1W4XIP5 Q16PI2 K7IRU0 A0A1Q3EZ22 A0A232FHG3 A0A154P0F8 A0A0T6AVV8 A0A0N0U5D8 A0A0L7R1B4 B0XHF9 A0A182NNB4 A0A182QXG2 E1ZZE0 A0A182WZV9 A0A182HXH3 A0A182LI76 A0A182P603 A7UUW8 A0A1S4GM06 Q7Q2P2 A0A182R2R0 A0A182VN66 A0A182UH59 A0A0C9Q2T3 A0A182JS38 A0A182MED6 A0A182WAQ9 A0A1B0GHL3 A0A1B6C294 A0A310SSV8 A0A182YI35 H3DEQ7 Q4RSS3 A0A1Y1LYS6 A0A2M4BFD2 A0A2M4BFD1 W5MPB8 A0A182SR75 H2UY09 M3XJD5 A0A2M4CS67 A0A0K8TMH3 A0A2M4CS99 H3AB96 A0A182FT65 H3AB95 H9GES1 K1R6Q2 A0A210Q6D9 K1PM37 A0A3Q3BXD0 A0A218UJ95 A0A2K6LJ63 A0A3B4GLP3 U3JX82 A0A3P8NDV3 A0A3P9CJJ3 A0A2D4NX59 A0A3Q4MC66 A0A2D4IW92 F7G201 A0A3B4GLR0 A0A2D4JZQ7 I3K7T4 T1E3X5 G3VHX2 A0A3M0K4R3 J3S191
A0A067QMF0 A0A2J7RRE6 F4WXM1 A0A151WSW5 A0A195FQK8 A0A026W386 A0A151JR84 A0A195BXH5 A0A195CRL6 A0A158NUX2 A0A336KZH7 A0A1B6MH52 A0A182JCL4 A0A336LT34 A0A1B6ENV1 D6WXN9 E2BKD1 E9ID99 A0A182G622 A0A1B0DAS6 A0A088AJP7 U5EKA3 A0A084VZT0 A0A2A3E2Y0 A0A1L8DCK4 A0A1W4XIP5 Q16PI2 K7IRU0 A0A1Q3EZ22 A0A232FHG3 A0A154P0F8 A0A0T6AVV8 A0A0N0U5D8 A0A0L7R1B4 B0XHF9 A0A182NNB4 A0A182QXG2 E1ZZE0 A0A182WZV9 A0A182HXH3 A0A182LI76 A0A182P603 A7UUW8 A0A1S4GM06 Q7Q2P2 A0A182R2R0 A0A182VN66 A0A182UH59 A0A0C9Q2T3 A0A182JS38 A0A182MED6 A0A182WAQ9 A0A1B0GHL3 A0A1B6C294 A0A310SSV8 A0A182YI35 H3DEQ7 Q4RSS3 A0A1Y1LYS6 A0A2M4BFD2 A0A2M4BFD1 W5MPB8 A0A182SR75 H2UY09 M3XJD5 A0A2M4CS67 A0A0K8TMH3 A0A2M4CS99 H3AB96 A0A182FT65 H3AB95 H9GES1 K1R6Q2 A0A210Q6D9 K1PM37 A0A3Q3BXD0 A0A218UJ95 A0A2K6LJ63 A0A3B4GLP3 U3JX82 A0A3P8NDV3 A0A3P9CJJ3 A0A2D4NX59 A0A3Q4MC66 A0A2D4IW92 F7G201 A0A3B4GLR0 A0A2D4JZQ7 I3K7T4 T1E3X5 G3VHX2 A0A3M0K4R3 J3S191
Pubmed
19121390
26354079
22118469
24845553
21719571
24508170
+ More
30249741 21347285 18362917 19820115 20798317 21282665 26483478 24438588 17510324 20075255 28648823 20966253 12364791 25244985 15496914 28004739 21551351 9215903 26369729 21881562 22992520 28812685 25186727 17495919 23758969 21709235 23025625
30249741 21347285 18362917 19820115 20798317 21282665 26483478 24438588 17510324 20075255 28648823 20966253 12364791 25244985 15496914 28004739 21551351 9215903 26369729 21881562 22992520 28812685 25186727 17495919 23758969 21709235 23025625
EMBL
BABH01013946
ODYU01004863
SOQ45178.1
RSAL01000041
RVE50855.1
KQ459595
+ More
KPI95595.1 KQ461154 KPJ08524.1 AGBW02011442 OWR46636.1 KK853247 KDR09377.1 NEVH01000610 PNF43385.1 GL888425 EGI61091.1 KQ982769 KYQ50928.1 KQ981335 KYN42582.1 KK107455 QOIP01000010 EZA50540.1 RLU17580.1 KQ978622 KYN29735.1 KQ976401 KYM92646.1 KQ977444 KYN02754.1 ADTU01026810 ADTU01026811 UFQS01000715 UFQT01000715 SSX06360.1 SSX26714.1 GEBQ01004697 JAT35280.1 UFQT01000099 SSX19893.1 GECZ01030158 JAS39611.1 KQ971362 EFA07957.1 GL448783 EFN83855.1 GL762454 EFZ21357.1 JXUM01147161 KQ570310 KXJ68389.1 AJVK01013388 GANO01001885 JAB57986.1 ATLV01018955 KE525256 KFB43474.1 KZ288409 PBC26103.1 GFDF01009969 JAV04115.1 CH477781 EAT36266.1 GFDL01014557 JAV20488.1 NNAY01000187 OXU30184.1 KQ434782 KZC04630.1 LJIG01022695 KRT79206.1 KQ435783 KOX74624.1 KQ414668 KOC64632.1 DS233154 EDS28388.1 AXCN02001904 GL435343 EFN73432.1 APCN01005305 AAAB01008968 EDO63511.1 EAA13229.4 GBYB01008318 JAG78085.1 AXCM01000075 AJWK01005316 GEDC01029662 JAS07636.1 KQ760688 OAD59358.1 CAAE01014999 CAG08559.1 GEZM01045195 JAV77868.1 GGFJ01002619 MBW51760.1 GGFJ01002618 MBW51759.1 AHAT01005282 AFYH01195898 AFYH01195899 AFYH01195900 AFYH01195901 AFYH01195902 AFYH01195903 GGFL01004018 MBW68196.1 GDAI01002508 JAI15095.1 GGFL01004019 MBW68197.1 AAWZ02027449 JH818316 EKC29636.1 NEDP02004827 OWF44303.1 JH815834 EKC19934.1 MUZQ01000290 OWK53442.1 AGTO01020421 IACN01036342 LAB50331.1 IACK01134014 LAA88530.1 IACL01025405 LAB01935.1 AERX01026869 GAAZ01002565 JAA95378.1 AEFK01093719 AEFK01093720 AEFK01093721 QRBI01000121 RMC06050.1 JU176161 GBEX01003503 AFJ51684.1 JAI11057.1
KPI95595.1 KQ461154 KPJ08524.1 AGBW02011442 OWR46636.1 KK853247 KDR09377.1 NEVH01000610 PNF43385.1 GL888425 EGI61091.1 KQ982769 KYQ50928.1 KQ981335 KYN42582.1 KK107455 QOIP01000010 EZA50540.1 RLU17580.1 KQ978622 KYN29735.1 KQ976401 KYM92646.1 KQ977444 KYN02754.1 ADTU01026810 ADTU01026811 UFQS01000715 UFQT01000715 SSX06360.1 SSX26714.1 GEBQ01004697 JAT35280.1 UFQT01000099 SSX19893.1 GECZ01030158 JAS39611.1 KQ971362 EFA07957.1 GL448783 EFN83855.1 GL762454 EFZ21357.1 JXUM01147161 KQ570310 KXJ68389.1 AJVK01013388 GANO01001885 JAB57986.1 ATLV01018955 KE525256 KFB43474.1 KZ288409 PBC26103.1 GFDF01009969 JAV04115.1 CH477781 EAT36266.1 GFDL01014557 JAV20488.1 NNAY01000187 OXU30184.1 KQ434782 KZC04630.1 LJIG01022695 KRT79206.1 KQ435783 KOX74624.1 KQ414668 KOC64632.1 DS233154 EDS28388.1 AXCN02001904 GL435343 EFN73432.1 APCN01005305 AAAB01008968 EDO63511.1 EAA13229.4 GBYB01008318 JAG78085.1 AXCM01000075 AJWK01005316 GEDC01029662 JAS07636.1 KQ760688 OAD59358.1 CAAE01014999 CAG08559.1 GEZM01045195 JAV77868.1 GGFJ01002619 MBW51760.1 GGFJ01002618 MBW51759.1 AHAT01005282 AFYH01195898 AFYH01195899 AFYH01195900 AFYH01195901 AFYH01195902 AFYH01195903 GGFL01004018 MBW68196.1 GDAI01002508 JAI15095.1 GGFL01004019 MBW68197.1 AAWZ02027449 JH818316 EKC29636.1 NEDP02004827 OWF44303.1 JH815834 EKC19934.1 MUZQ01000290 OWK53442.1 AGTO01020421 IACN01036342 LAB50331.1 IACK01134014 LAA88530.1 IACL01025405 LAB01935.1 AERX01026869 GAAZ01002565 JAA95378.1 AEFK01093719 AEFK01093720 AEFK01093721 QRBI01000121 RMC06050.1 JU176161 GBEX01003503 AFJ51684.1 JAI11057.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000007151
UP000027135
+ More
UP000235965 UP000007755 UP000075809 UP000078541 UP000053097 UP000279307 UP000078492 UP000078540 UP000078542 UP000005205 UP000075880 UP000007266 UP000008237 UP000069940 UP000249989 UP000092462 UP000005203 UP000030765 UP000242457 UP000192223 UP000008820 UP000002358 UP000215335 UP000076502 UP000053105 UP000053825 UP000002320 UP000075884 UP000075886 UP000000311 UP000076407 UP000075840 UP000075882 UP000075885 UP000007062 UP000075900 UP000075903 UP000075902 UP000075881 UP000075883 UP000075920 UP000092461 UP000076408 UP000007303 UP000018468 UP000075901 UP000005226 UP000008672 UP000069272 UP000001646 UP000005408 UP000242188 UP000264840 UP000197619 UP000233180 UP000261460 UP000016665 UP000265100 UP000265160 UP000261580 UP000002280 UP000005207 UP000007648 UP000269221
UP000235965 UP000007755 UP000075809 UP000078541 UP000053097 UP000279307 UP000078492 UP000078540 UP000078542 UP000005205 UP000075880 UP000007266 UP000008237 UP000069940 UP000249989 UP000092462 UP000005203 UP000030765 UP000242457 UP000192223 UP000008820 UP000002358 UP000215335 UP000076502 UP000053105 UP000053825 UP000002320 UP000075884 UP000075886 UP000000311 UP000076407 UP000075840 UP000075882 UP000075885 UP000007062 UP000075900 UP000075903 UP000075902 UP000075881 UP000075883 UP000075920 UP000092461 UP000076408 UP000007303 UP000018468 UP000075901 UP000005226 UP000008672 UP000069272 UP000001646 UP000005408 UP000242188 UP000264840 UP000197619 UP000233180 UP000261460 UP000016665 UP000265100 UP000265160 UP000261580 UP000002280 UP000005207 UP000007648 UP000269221
PRIDE
ProteinModelPortal
H9J725
A0A2H1VWJ8
A0A3S2PGM4
A0A194PWT4
A0A194QT81
A0A212EYR7
+ More
A0A067QMF0 A0A2J7RRE6 F4WXM1 A0A151WSW5 A0A195FQK8 A0A026W386 A0A151JR84 A0A195BXH5 A0A195CRL6 A0A158NUX2 A0A336KZH7 A0A1B6MH52 A0A182JCL4 A0A336LT34 A0A1B6ENV1 D6WXN9 E2BKD1 E9ID99 A0A182G622 A0A1B0DAS6 A0A088AJP7 U5EKA3 A0A084VZT0 A0A2A3E2Y0 A0A1L8DCK4 A0A1W4XIP5 Q16PI2 K7IRU0 A0A1Q3EZ22 A0A232FHG3 A0A154P0F8 A0A0T6AVV8 A0A0N0U5D8 A0A0L7R1B4 B0XHF9 A0A182NNB4 A0A182QXG2 E1ZZE0 A0A182WZV9 A0A182HXH3 A0A182LI76 A0A182P603 A7UUW8 A0A1S4GM06 Q7Q2P2 A0A182R2R0 A0A182VN66 A0A182UH59 A0A0C9Q2T3 A0A182JS38 A0A182MED6 A0A182WAQ9 A0A1B0GHL3 A0A1B6C294 A0A310SSV8 A0A182YI35 H3DEQ7 Q4RSS3 A0A1Y1LYS6 A0A2M4BFD2 A0A2M4BFD1 W5MPB8 A0A182SR75 H2UY09 M3XJD5 A0A2M4CS67 A0A0K8TMH3 A0A2M4CS99 H3AB96 A0A182FT65 H3AB95 H9GES1 K1R6Q2 A0A210Q6D9 K1PM37 A0A3Q3BXD0 A0A218UJ95 A0A2K6LJ63 A0A3B4GLP3 U3JX82 A0A3P8NDV3 A0A3P9CJJ3 A0A2D4NX59 A0A3Q4MC66 A0A2D4IW92 F7G201 A0A3B4GLR0 A0A2D4JZQ7 I3K7T4 T1E3X5 G3VHX2 A0A3M0K4R3 J3S191
A0A067QMF0 A0A2J7RRE6 F4WXM1 A0A151WSW5 A0A195FQK8 A0A026W386 A0A151JR84 A0A195BXH5 A0A195CRL6 A0A158NUX2 A0A336KZH7 A0A1B6MH52 A0A182JCL4 A0A336LT34 A0A1B6ENV1 D6WXN9 E2BKD1 E9ID99 A0A182G622 A0A1B0DAS6 A0A088AJP7 U5EKA3 A0A084VZT0 A0A2A3E2Y0 A0A1L8DCK4 A0A1W4XIP5 Q16PI2 K7IRU0 A0A1Q3EZ22 A0A232FHG3 A0A154P0F8 A0A0T6AVV8 A0A0N0U5D8 A0A0L7R1B4 B0XHF9 A0A182NNB4 A0A182QXG2 E1ZZE0 A0A182WZV9 A0A182HXH3 A0A182LI76 A0A182P603 A7UUW8 A0A1S4GM06 Q7Q2P2 A0A182R2R0 A0A182VN66 A0A182UH59 A0A0C9Q2T3 A0A182JS38 A0A182MED6 A0A182WAQ9 A0A1B0GHL3 A0A1B6C294 A0A310SSV8 A0A182YI35 H3DEQ7 Q4RSS3 A0A1Y1LYS6 A0A2M4BFD2 A0A2M4BFD1 W5MPB8 A0A182SR75 H2UY09 M3XJD5 A0A2M4CS67 A0A0K8TMH3 A0A2M4CS99 H3AB96 A0A182FT65 H3AB95 H9GES1 K1R6Q2 A0A210Q6D9 K1PM37 A0A3Q3BXD0 A0A218UJ95 A0A2K6LJ63 A0A3B4GLP3 U3JX82 A0A3P8NDV3 A0A3P9CJJ3 A0A2D4NX59 A0A3Q4MC66 A0A2D4IW92 F7G201 A0A3B4GLR0 A0A2D4JZQ7 I3K7T4 T1E3X5 G3VHX2 A0A3M0K4R3 J3S191
Ontologies
GO
PANTHER
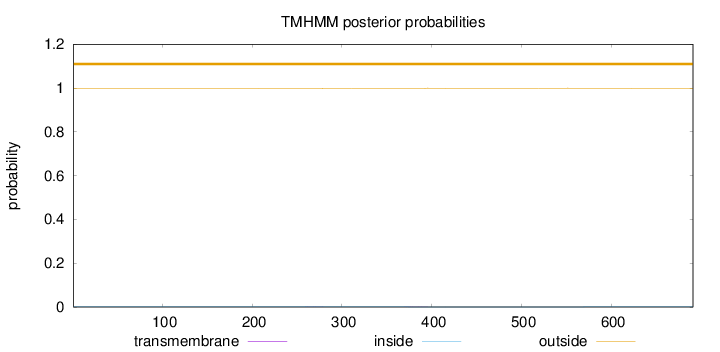
Topology
Length:
691
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02987
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00155
outside
1 - 691
Population Genetic Test Statistics
Pi
257.154372
Theta
140.578695
Tajima's D
-0.914944
CLR
321.564278
CSRT
0.155792210389481
Interpretation
Possibly Positive selection
