Gene
KWMTBOMO04603
Pre Gene Modal
BGIBMGA005492
Annotation
ATG1_transcript_variant_A_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.505
Sequence
CDS
ATGCTTAGTAATATTTTACGCTTGAAGTTTGACACAACAGATTACTGCAATGGAGGCGACCTAGCGGATTATCTTCAAACGAACCGGCTGCTCAGCGAGACCACGATCCAGTTGTTCTTAGCGCAGCTTGCAGAAGCGATGAGCGCCATCCACGCGAAGGGCATCGTCCACCGTGACTTGAAGCCGCAAAACATACTCCTCACTCATTCTATCCTACCGCCTCGTACTCCACACCCATCAGATATAACACTCAAAATTGCTGACTTCGGATTCGCAAGGTTCCTGGAAGAAGGAAATATGGCCGTTACGCTCTGCGGCTCCCCAATGTACATGGCTCCAGAAGTCATAATGTCCCTGAAATACGACGCCAAAGCAGACCTGTGGAGCCTCGGCACAATAGTCTACCAGTGCCTTACCGGGAAGGCTCCCTTCCAAGCCACCACGCCACACGAACTGAAAGCTTTCTATGAAAATAGCGTCGATCTTCAGCCCAAAATCCCACCAGGAACGAGTCCAGAACTGTGCAGCTTATTGATTGGCCTACTTCGACGAAACCCTCGAGAACGCATGTCATTCGAAATGTTCTTCAATCATCCCTTTCTTCAACGATCGCGCAACACAAACGTCCCCAAGATGGGATCGTGTGCGGGTCCGTCCGTGGTGGCGAACCGAGCGCCCCCCTCCCGCGCCACCCCGCCCGCGCCCCTGCCCCTAGCCTTACCCACTGCCAAGAAAACACAGCCAACATCCTCCGATTCGTCGGAGGTTGGTTGGGCGGGTGAAGAGGACTTTGTGTTGGTGGAGGCGACGGAGAGCGGGTCCGCAGAGTGCTCGGCCGCCTCCTCCGGCTCGGAGGCAGCGTCCAGACCCCGCTCCCTGGCGCTCCCCGCGCCCCCCGCACCCGCCTCCCCTTGCACACCTACCGCGAGCCCACTGACCGCGGCCACTAACACTACTGTACCGCGATCTCAGCCCATCGACGTGCGCCGAACGACCTACAGCCGCAACGCCACGAACATCGGCTCGCTCTCGCCGCCCACTATGCCGTTCGCGATGGGCACGCCGCCGTCGACGCGGCGTCGCAGCAGCAACAGCGGCAGCTCCCCGCCGCCGCAGCTGTGGCAGGTGTCGCCCACCAACGCGAGCCCGCTGCGCAGGTCCGGCTGGCCGGCCGCAGGCTCGTCCCCGCCCAGTGCGCTGGCGCTGCGTGCGAGCGGGGAGTGCGGGGGCGGCGCGGGGGGCGCGCTCGGCGACGTGCTGCGCGGCATGAAGCCCGACCCGCCCGTCTACATACCCAACCTGCAGGAGGAGACCATACTCGCGGAGGAACACAGCAAAGTGTCATCACAACTTAACTTCGTTCTGATGTTGGCCGAACTGCTGTCGGACTTGGCCGTTTCTTGCGGAGCTCCCATCGCAGCCCTGATGGATACGTCCGATGAGCGTTGCAACGAGAGCGTCCGGCTGGGGCTGTTGGTGCAGGCCATGCAGGCGCTCGCAGCGGGCCTGCGCCTGGCCGCCACGCACTACCGCGCGCGCACGCTGCAGCCCACGCCGCAAGTGCGCAGCGTTGTATCCCTGATGAACGGTAAATACAAATGGATCGTCAACGAGTCGAAGCGACTGCACGAAGCGGGAGTGACGCCAGCCGTCTGCGATAAAATACTATACGAGCACGCTATTGAACTGTGTCAGATGGCGGCCATCGAGGAACTGTTCGGCGACACGAAGGAGTGCGAGCGCCGCTACATATCGGCCCAGGTGCTGCTGCACTCGCTCGTGCAGAGACACCCGCTGCATCCACACCACCGGACCACGCTCTCCAAATATCGAGACGCAGTACAACGTAGACTAAACTGTCTCAAAGGTGTTCGCAAAATAATGGATGTCAAAATTGAAGCCGGGATCTCATAA
Protein
MLSNILRLKFDTTDYCNGGDLADYLQTNRLLSETTIQLFLAQLAEAMSAIHAKGIVHRDLKPQNILLTHSILPPRTPHPSDITLKIADFGFARFLEEGNMAVTLCGSPMYMAPEVIMSLKYDAKADLWSLGTIVYQCLTGKAPFQATTPHELKAFYENSVDLQPKIPPGTSPELCSLLIGLLRRNPRERMSFEMFFNHPFLQRSRNTNVPKMGSCAGPSVVANRAPPSRATPPAPLPLALPTAKKTQPTSSDSSEVGWAGEEDFVLVEATESGSAECSAASSGSEAASRPRSLALPAPPAPASPCTPTASPLTAATNTTVPRSQPIDVRRTTYSRNATNIGSLSPPTMPFAMGTPPSTRRRSSNSGSSPPPQLWQVSPTNASPLRRSGWPAAGSSPPSALALRASGECGGGAGGALGDVLRGMKPDPPVYIPNLQEETILAEEHSKVSSQLNFVLMLAELLSDLAVSCGAPIAALMDTSDERCNESVRLGLLVQAMQALAAGLRLAATHYRARTLQPTPQVRSVVSLMNGKYKWIVNESKRLHEAGVTPAVCDKILYEHAIELCQMAAIEELFGDTKECERRYISAQVLLHSLVQRHPLHPHHRTTLSKYRDAVQRRLNCLKGVRKIMDVKIEAGIS
Summary
Uniprot
Pubmed
EMBL
Proteomes
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
Ontologies
PATHWAY
GO
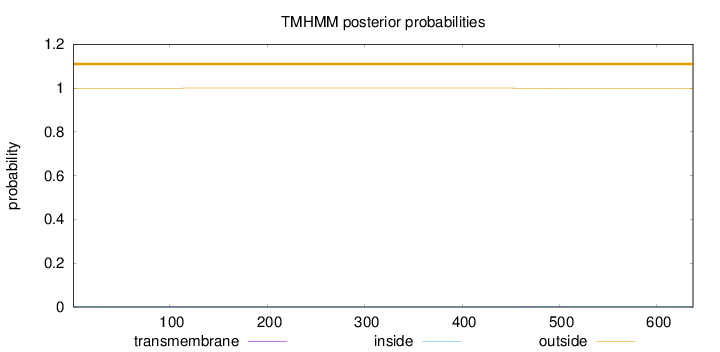
Topology
Length:
637
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04578
Exp number, first 60 AAs:
0.00213
Total prob of N-in:
0.00060
outside
1 - 637
Population Genetic Test Statistics
Pi
194.788857
Theta
160.211982
Tajima's D
-0.798522
CLR
256.831079
CSRT
0.180040997950103
Interpretation
Uncertain
