Gene
KWMTBOMO04598
Pre Gene Modal
BGIBMGA005490
Annotation
PREDICTED:_septin-2_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 4.34
Sequence
CDS
ATGGAAAACAGTGATTCTTTCGCTATATTAGACATTAATCTCAGTGATAAGGATAAGGACGAACTGAAAAAAAATGATGGAAATAAAAAATCGCCAATCACAACTGTCATAAAAATTCAACGTGATCGCGGTGAACGCGACTACATTGGATTTGCTACATTGCCTGAACAGGTGCACCGCAAATCAGTGAAGAGAGGTTTCGACTTCACTCTCATGGTTGTCGGAGAATCAGGCCTCGGAAAATCAACATTGATAAACAGTTTATTTTTAGGAGATCTTTACAAAAATCGAAAAATTCCAGATGTTCAAGATAGAATAGAAAAAACGACGACAATCGAAAAGAAAACTATGGAAATAGAAGAACGAGGCGTCAAATTACGTCTGACGATTGTTGACACGCCTGGCTTTGGAGACGCGATTAACTGTGAAGACTCGTGGCGCGTTTGTTCAGCATACATTGACGAACAATTCAGGCAGTATTTTACTGATGAAAGTGGTCTGAACAGGCGACATATGCAGGACAACCGAGTTCATTGCTGTCTTTACTTCGTTCCGCCTTGGGCGCATAGCCTACGACAAGTGGATCTCGAAATGATGAAGCGTCTGCATCGCAAAGTGAACATCGTGGTGGTAATCGCAAAAGCGGACTCTCTTACGGCGATCGAAATAAAACGACTCAAGGCACGAATACTTAACGATCTTGAAGAACACCAGATACAAGTATACCAATTCCCGGAATGTGACAGCGACGAAGATGAAGACTTCAAACAACAAGACCGCGAGTTAAAGGAAGCGGCCCCATTTGCTGTGGTCGCTTCTGATATAGTCCTCGAGATGGGAGGCAAAAGAGTGCGGGGTAGACAGTATCCGTGGGGGATTGTTGATGTTGAAAATCCACGGCATTCAGACTTCACCAAACTTAGAACAATGTTAATTTCTACACACATGCAGGATCTGAAGGACGTGACTCAAGATGTGCACTACGAAAATTTCCGTGCTCAGTGCATTTCTCAAATTTCACAACATGCCATGCGTGAACGGGGGAAACTGAAGAGAGACTCTATGGGTAACAACAACGACGTGGTCATCACCGACACTGATCGACTCTTGCTCCAAAAAGACGAAGAGATACGAAGAATGCAAGATATGTTAACTCAAATGCAACAAAAACTAAAAGCTTCCGATAAAAAACACGACAGTATAATAGATGTATAG
Protein
MENSDSFAILDINLSDKDKDELKKNDGNKKSPITTVIKIQRDRGERDYIGFATLPEQVHRKSVKRGFDFTLMVVGESGLGKSTLINSLFLGDLYKNRKIPDVQDRIEKTTTIEKKTMEIEERGVKLRLTIVDTPGFGDAINCEDSWRVCSAYIDEQFRQYFTDESGLNRRHMQDNRVHCCLYFVPPWAHSLRQVDLEMMKRLHRKVNIVVVIAKADSLTAIEIKRLKARILNDLEEHQIQVYQFPECDSDEDEDFKQQDRELKEAAPFAVVASDIVLEMGGKRVRGRQYPWGIVDVENPRHSDFTKLRTMLISTHMQDLKDVTQDVHYENFRAQCISQISQHAMRERGKLKRDSMGNNNDVVITDTDRLLLQKDEEIRRMQDMLTQMQQKLKASDKKHDSIIDV
Summary
Similarity
Belongs to the TRAFAC class TrmE-Era-EngA-EngB-Septin-like GTPase superfamily. Septin GTPase family.
Belongs to the FBPase class 1 family.
Belongs to the FBPase class 1 family.
Uniprot
A0A2H1V3E0
H9J7J8
A0A212FJU8
A0A194PS60
A0A194QSL7
A0A1B0DJT3
+ More
A0A1B0CR21 A0A1Y1KDK9 W8C491 A0A1Y1KII0 A0A1A9VB19 A0A1B0BR17 A0A0A1XN48 A0A0K8W6W5 W8BIJ7 B4NCS1 A0A034VYS3 A0A1I8MBF4 A0A3B0K4R1 B3MWA1 A0A0Q5TJB7 B3NVL0 N6U4Q6 Q0KHR7 A0A1I8PED1 U4ULY1 A0A3B0JXG6 Q29HN6 A0A0R3P4M9 A0A1W4VWP2 B4R611 A0A0R1EBV8 Q7KUX3 B4Q1T4 B4JN03 A0A1W4VVW0 A0A1A9Y2W4 A0A0M3QZE1 Q960I4 A0A1I8PED8 A0A2P8YLB5 A0A0L0C9Q1 B4MCU4 A0A1W4X3P3 C8VV45 A0A146M6Q7 A0A0A9Y058 A0A023F711 J3JX28 D6WXZ1 T1IFQ6 E2B2Z9 A0A067QQ62 A0A0L7R770 F4W8S0 A0A026X2S1 A0A0A9Y2S3 A0A195EIH3 A0A158NKG5 A0A1B6GTR9 V9IGR0 E1ZZZ1 E0VTS6 A0A336MBH5 A0A1B6BYA2 A0A182J432 A0A084VV68 A0A023EQ06 A0A1S4FS27 Q16RA7 A0A1Q3FCX6 B0WZY0 A0A1Q3FDM0 A0A1Q3FE75 A0A1S4G9T6 A0A182UN23 A0A182LIM0 A0A182XEK7 Q7QE93 A0A2M4AG18 A0A195B034 A0A182FMF8 A0A182QM99 A0A151WT59 W5JKM8 A0A2M4BST8 A0A182U289 A0A182I9J5 A0A0A1XLH6 A0A0K8UMM6 A0A182P6U4 A0A1B0FGR1 A0A151IJX6 T1PJV1 A0A3L8DB26 A0A1W4XE38 A0A182MML4 A0A0P4VWL8 Q1DH20 A0A182W470
A0A1B0CR21 A0A1Y1KDK9 W8C491 A0A1Y1KII0 A0A1A9VB19 A0A1B0BR17 A0A0A1XN48 A0A0K8W6W5 W8BIJ7 B4NCS1 A0A034VYS3 A0A1I8MBF4 A0A3B0K4R1 B3MWA1 A0A0Q5TJB7 B3NVL0 N6U4Q6 Q0KHR7 A0A1I8PED1 U4ULY1 A0A3B0JXG6 Q29HN6 A0A0R3P4M9 A0A1W4VWP2 B4R611 A0A0R1EBV8 Q7KUX3 B4Q1T4 B4JN03 A0A1W4VVW0 A0A1A9Y2W4 A0A0M3QZE1 Q960I4 A0A1I8PED8 A0A2P8YLB5 A0A0L0C9Q1 B4MCU4 A0A1W4X3P3 C8VV45 A0A146M6Q7 A0A0A9Y058 A0A023F711 J3JX28 D6WXZ1 T1IFQ6 E2B2Z9 A0A067QQ62 A0A0L7R770 F4W8S0 A0A026X2S1 A0A0A9Y2S3 A0A195EIH3 A0A158NKG5 A0A1B6GTR9 V9IGR0 E1ZZZ1 E0VTS6 A0A336MBH5 A0A1B6BYA2 A0A182J432 A0A084VV68 A0A023EQ06 A0A1S4FS27 Q16RA7 A0A1Q3FCX6 B0WZY0 A0A1Q3FDM0 A0A1Q3FE75 A0A1S4G9T6 A0A182UN23 A0A182LIM0 A0A182XEK7 Q7QE93 A0A2M4AG18 A0A195B034 A0A182FMF8 A0A182QM99 A0A151WT59 W5JKM8 A0A2M4BST8 A0A182U289 A0A182I9J5 A0A0A1XLH6 A0A0K8UMM6 A0A182P6U4 A0A1B0FGR1 A0A151IJX6 T1PJV1 A0A3L8DB26 A0A1W4XE38 A0A182MML4 A0A0P4VWL8 Q1DH20 A0A182W470
Pubmed
19121390
22118469
26354079
28004739
24495485
25830018
+ More
17994087 25348373 25315136 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 29403074 26108605 26823975 25401762 25474469 22516182 18362917 19820115 20798317 24845553 21719571 24508170 21347285 20566863 24438588 24945155 17510324 12364791 20966253 20920257 23761445 30249741
17994087 25348373 25315136 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 29403074 26108605 26823975 25401762 25474469 22516182 18362917 19820115 20798317 24845553 21719571 24508170 21347285 20566863 24438588 24945155 17510324 12364791 20966253 20920257 23761445 30249741
EMBL
ODYU01000492
SOQ35360.1
BABH01013940
AGBW02008208
OWR54004.1
KQ459595
+ More
KPI95589.1 KQ461154 KPJ08518.1 AJVK01001049 AJWK01024346 GEZM01087601 JAV58588.1 GAMC01009716 JAB96839.1 GEZM01087602 JAV58587.1 JXJN01018870 GBXI01001856 JAD12436.1 GDHF01005684 JAI46630.1 GAMC01009717 JAB96838.1 CH964239 EDW82630.2 GAKP01011912 JAC47040.1 OUUW01000003 SPP78458.1 CH902625 EDV35246.2 CH954180 KQS29962.1 EDV46402.1 APGK01040123 KB740975 ENN76565.1 AE014298 AAF48645.1 AAF48646.1 AAN09414.1 AAN09415.1 AAN09416.1 AAN09417.1 KB632267 ERL91010.1 SPP78459.1 CH379064 EAL31722.3 KRT06485.1 CM000366 EDX18115.1 CM000162 KRK06809.1 BT132948 AAS65390.1 AEW43888.1 EDX02509.1 KRK06807.1 KRK06808.1 CH916371 EDV92096.1 CP012528 ALC49263.1 AY052045 AAK93469.1 PYGN01000512 PSN45049.1 JRES01000728 KNC28935.1 CH940660 EDW58016.2 BT099791 ACV91628.1 GDHC01003191 JAQ15438.1 GBHO01017152 JAG26452.1 GBBI01001687 JAC17025.1 BT127796 AEE62758.1 KQ971362 EFA08920.1 ACPB03005594 GL445250 EFN89968.1 KK853603 KDR06354.1 KQ414642 KOC66727.1 GL887908 EGI69561.1 KK107021 EZA62393.1 GBHO01017150 JAG26454.1 KQ978881 KYN27697.1 ADTU01018831 GECZ01003974 JAS65795.1 JR046791 AEY60250.1 GL435531 EFN73231.1 DS235773 EEB16782.1 UFQT01000707 SSX26681.1 GEDC01031062 JAS06236.1 ATLV01017157 KE525157 KFB41862.1 GAPW01001966 JAC11632.1 CH477719 EAT36940.1 GFDL01009594 JAV25451.1 DS232221 EDS37805.1 GFDL01009436 JAV25609.1 GFDL01009237 JAV25808.1 AAAB01008847 EAA06877.5 GGFK01006413 MBW39734.1 KQ976692 KYM77838.1 AXCN02001412 KQ982762 KYQ51070.1 ADMH02001263 ETN63334.1 GGFJ01006850 MBW55991.1 APCN01000987 GBXI01006144 GBXI01002471 JAD08148.1 JAD11821.1 GDHF01024689 JAI27625.1 CCAG010020401 KQ977294 KYN03951.1 KA649036 AFP63665.1 QOIP01000011 RLU17079.1 AXCM01001219 GDRN01091225 JAI60314.1 CH899827 EAT32450.1
KPI95589.1 KQ461154 KPJ08518.1 AJVK01001049 AJWK01024346 GEZM01087601 JAV58588.1 GAMC01009716 JAB96839.1 GEZM01087602 JAV58587.1 JXJN01018870 GBXI01001856 JAD12436.1 GDHF01005684 JAI46630.1 GAMC01009717 JAB96838.1 CH964239 EDW82630.2 GAKP01011912 JAC47040.1 OUUW01000003 SPP78458.1 CH902625 EDV35246.2 CH954180 KQS29962.1 EDV46402.1 APGK01040123 KB740975 ENN76565.1 AE014298 AAF48645.1 AAF48646.1 AAN09414.1 AAN09415.1 AAN09416.1 AAN09417.1 KB632267 ERL91010.1 SPP78459.1 CH379064 EAL31722.3 KRT06485.1 CM000366 EDX18115.1 CM000162 KRK06809.1 BT132948 AAS65390.1 AEW43888.1 EDX02509.1 KRK06807.1 KRK06808.1 CH916371 EDV92096.1 CP012528 ALC49263.1 AY052045 AAK93469.1 PYGN01000512 PSN45049.1 JRES01000728 KNC28935.1 CH940660 EDW58016.2 BT099791 ACV91628.1 GDHC01003191 JAQ15438.1 GBHO01017152 JAG26452.1 GBBI01001687 JAC17025.1 BT127796 AEE62758.1 KQ971362 EFA08920.1 ACPB03005594 GL445250 EFN89968.1 KK853603 KDR06354.1 KQ414642 KOC66727.1 GL887908 EGI69561.1 KK107021 EZA62393.1 GBHO01017150 JAG26454.1 KQ978881 KYN27697.1 ADTU01018831 GECZ01003974 JAS65795.1 JR046791 AEY60250.1 GL435531 EFN73231.1 DS235773 EEB16782.1 UFQT01000707 SSX26681.1 GEDC01031062 JAS06236.1 ATLV01017157 KE525157 KFB41862.1 GAPW01001966 JAC11632.1 CH477719 EAT36940.1 GFDL01009594 JAV25451.1 DS232221 EDS37805.1 GFDL01009436 JAV25609.1 GFDL01009237 JAV25808.1 AAAB01008847 EAA06877.5 GGFK01006413 MBW39734.1 KQ976692 KYM77838.1 AXCN02001412 KQ982762 KYQ51070.1 ADMH02001263 ETN63334.1 GGFJ01006850 MBW55991.1 APCN01000987 GBXI01006144 GBXI01002471 JAD08148.1 JAD11821.1 GDHF01024689 JAI27625.1 CCAG010020401 KQ977294 KYN03951.1 KA649036 AFP63665.1 QOIP01000011 RLU17079.1 AXCM01001219 GDRN01091225 JAI60314.1 CH899827 EAT32450.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000092462
UP000092461
+ More
UP000078200 UP000092460 UP000007798 UP000095301 UP000268350 UP000007801 UP000008711 UP000019118 UP000000803 UP000095300 UP000030742 UP000001819 UP000192221 UP000000304 UP000002282 UP000001070 UP000092443 UP000092553 UP000245037 UP000037069 UP000008792 UP000192223 UP000007266 UP000015103 UP000008237 UP000027135 UP000053825 UP000007755 UP000053097 UP000078492 UP000005205 UP000000311 UP000009046 UP000075880 UP000030765 UP000008820 UP000002320 UP000075903 UP000075882 UP000076407 UP000007062 UP000078540 UP000069272 UP000075886 UP000075809 UP000000673 UP000075902 UP000075840 UP000075885 UP000092444 UP000078542 UP000279307 UP000075883 UP000075920
UP000078200 UP000092460 UP000007798 UP000095301 UP000268350 UP000007801 UP000008711 UP000019118 UP000000803 UP000095300 UP000030742 UP000001819 UP000192221 UP000000304 UP000002282 UP000001070 UP000092443 UP000092553 UP000245037 UP000037069 UP000008792 UP000192223 UP000007266 UP000015103 UP000008237 UP000027135 UP000053825 UP000007755 UP000053097 UP000078492 UP000005205 UP000000311 UP000009046 UP000075880 UP000030765 UP000008820 UP000002320 UP000075903 UP000075882 UP000076407 UP000007062 UP000078540 UP000069272 UP000075886 UP000075809 UP000000673 UP000075902 UP000075840 UP000075885 UP000092444 UP000078542 UP000279307 UP000075883 UP000075920
Interpro
Gene 3D
ProteinModelPortal
A0A2H1V3E0
H9J7J8
A0A212FJU8
A0A194PS60
A0A194QSL7
A0A1B0DJT3
+ More
A0A1B0CR21 A0A1Y1KDK9 W8C491 A0A1Y1KII0 A0A1A9VB19 A0A1B0BR17 A0A0A1XN48 A0A0K8W6W5 W8BIJ7 B4NCS1 A0A034VYS3 A0A1I8MBF4 A0A3B0K4R1 B3MWA1 A0A0Q5TJB7 B3NVL0 N6U4Q6 Q0KHR7 A0A1I8PED1 U4ULY1 A0A3B0JXG6 Q29HN6 A0A0R3P4M9 A0A1W4VWP2 B4R611 A0A0R1EBV8 Q7KUX3 B4Q1T4 B4JN03 A0A1W4VVW0 A0A1A9Y2W4 A0A0M3QZE1 Q960I4 A0A1I8PED8 A0A2P8YLB5 A0A0L0C9Q1 B4MCU4 A0A1W4X3P3 C8VV45 A0A146M6Q7 A0A0A9Y058 A0A023F711 J3JX28 D6WXZ1 T1IFQ6 E2B2Z9 A0A067QQ62 A0A0L7R770 F4W8S0 A0A026X2S1 A0A0A9Y2S3 A0A195EIH3 A0A158NKG5 A0A1B6GTR9 V9IGR0 E1ZZZ1 E0VTS6 A0A336MBH5 A0A1B6BYA2 A0A182J432 A0A084VV68 A0A023EQ06 A0A1S4FS27 Q16RA7 A0A1Q3FCX6 B0WZY0 A0A1Q3FDM0 A0A1Q3FE75 A0A1S4G9T6 A0A182UN23 A0A182LIM0 A0A182XEK7 Q7QE93 A0A2M4AG18 A0A195B034 A0A182FMF8 A0A182QM99 A0A151WT59 W5JKM8 A0A2M4BST8 A0A182U289 A0A182I9J5 A0A0A1XLH6 A0A0K8UMM6 A0A182P6U4 A0A1B0FGR1 A0A151IJX6 T1PJV1 A0A3L8DB26 A0A1W4XE38 A0A182MML4 A0A0P4VWL8 Q1DH20 A0A182W470
A0A1B0CR21 A0A1Y1KDK9 W8C491 A0A1Y1KII0 A0A1A9VB19 A0A1B0BR17 A0A0A1XN48 A0A0K8W6W5 W8BIJ7 B4NCS1 A0A034VYS3 A0A1I8MBF4 A0A3B0K4R1 B3MWA1 A0A0Q5TJB7 B3NVL0 N6U4Q6 Q0KHR7 A0A1I8PED1 U4ULY1 A0A3B0JXG6 Q29HN6 A0A0R3P4M9 A0A1W4VWP2 B4R611 A0A0R1EBV8 Q7KUX3 B4Q1T4 B4JN03 A0A1W4VVW0 A0A1A9Y2W4 A0A0M3QZE1 Q960I4 A0A1I8PED8 A0A2P8YLB5 A0A0L0C9Q1 B4MCU4 A0A1W4X3P3 C8VV45 A0A146M6Q7 A0A0A9Y058 A0A023F711 J3JX28 D6WXZ1 T1IFQ6 E2B2Z9 A0A067QQ62 A0A0L7R770 F4W8S0 A0A026X2S1 A0A0A9Y2S3 A0A195EIH3 A0A158NKG5 A0A1B6GTR9 V9IGR0 E1ZZZ1 E0VTS6 A0A336MBH5 A0A1B6BYA2 A0A182J432 A0A084VV68 A0A023EQ06 A0A1S4FS27 Q16RA7 A0A1Q3FCX6 B0WZY0 A0A1Q3FDM0 A0A1Q3FE75 A0A1S4G9T6 A0A182UN23 A0A182LIM0 A0A182XEK7 Q7QE93 A0A2M4AG18 A0A195B034 A0A182FMF8 A0A182QM99 A0A151WT59 W5JKM8 A0A2M4BST8 A0A182U289 A0A182I9J5 A0A0A1XLH6 A0A0K8UMM6 A0A182P6U4 A0A1B0FGR1 A0A151IJX6 T1PJV1 A0A3L8DB26 A0A1W4XE38 A0A182MML4 A0A0P4VWL8 Q1DH20 A0A182W470
PDB
2QAG
E-value=6.42405e-108,
Score=999
Ontologies
GO
PANTHER
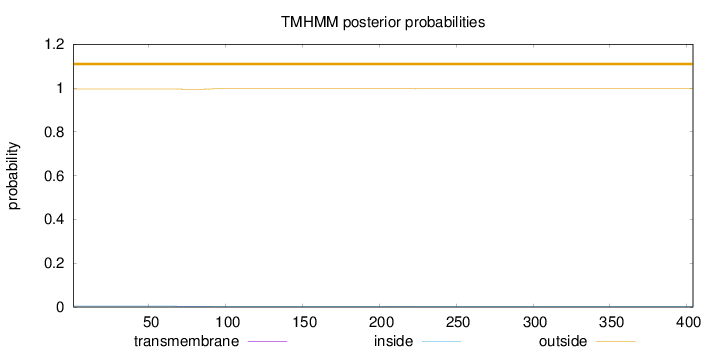
Topology
Length:
404
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0545
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00422
outside
1 - 404
Population Genetic Test Statistics
Pi
213.764105
Theta
175.246248
Tajima's D
0.584513
CLR
0.664805
CSRT
0.542722863856807
Interpretation
Uncertain
