Gene
KWMTBOMO04597 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005320
Annotation
PREDICTED:_carboxypeptidase_D-like_isoform_X4_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.436 Extracellular Reliability : 1.14
Sequence
CDS
ATGCTGGCTGGTTCGACATTTGCGCAATCTTTTCATAATCCAAGCTTAAATGGAGAAGAAACGTCAACCAATCTAGGTGAAGTGGAATCACGTGAAAATGCTCCAGACTTGGAGTTTCAATATCACGATCATGATCAGATGACGCGGTATCTACGTGCAGTTTCTGCCAAATATCCTGAACTCACAGCTTTATATTCCATTGGGAAGTCGGTACAAGGTCGAGATCTATGGGTGATGGTCGTCTCTGTGTCTCCATACGAGCACATGATTGGGAAGCCTGATGTTAAATATGTGGCTAACATACACGGTAACGAAGCAGTAGGACGGGAGCTACTTCTACATCTTATACAGTATTTAGTGACGTCGTATGCAACTGATCCGTACGTGAAATGGCTGTTGGACAACACACGTATTCACTTAATGCCCTCAATGAATCCTGATGGTTTCGCAATATCGAGGGAAGGACAATGCGACACCATACATGGCAGGCACAATGCACGTCGTCATGATTTGAACAGGAATTTTCCCGATTTTTTCAAAGCGAATACAAAACAGCCTCAGCCAGAAACGGAAGCAGTGAAACAATGGATCAGCAAAATACAGTTCGTTCTCTCTGGGTCGCTGCATGGCGGTGCTCTGGTCGCGTCTTATCCTTTCGACAACACTCCCAGTTCTACAGTGTTTCAAAACTATGAACACACGCCGTCAATATCTCCCGATGATGATGTCTTCAAACATTTGTCCTTGGTATACTCTAAAAACCATGCCAAAATGTCTCGAGGTGTCTCTTGCAAATCGGGATCACCAAGATTCGAAAATGGCATTACAAATGGAGCGGCCTGGTATCCACTGACAGGTGGCATGCAAGACTACAACTACCTGTGGCACGGATGTATGGAAGTCACTTTAGAAATATCTTGCTGTAAATATCCCCCAGCCCACGAATTACCCAAATACTGGCAAGACAATAAACAGTCTCTTATAAAATACTTAGCGGAAGTACACCGAGGTGCCCACGGCTTTGTCATGGATGAAAATAATAATCCAGTAGAGCGTGCAGCTATCAAAATCAAAGGACGAGATGTCAATTATCATACTACTAACTTCGGAGAATTTTGGCGCATTCTTTTACCCGGAACGTACCGCATGGACATTTCGGCGGAGGGATATTTACCACAAGAAGTAGAATTTATTGTTATTGATAGCCATCCAACGCTATTAAATGTTACTTTGCGTTCCGCGAAGGATGTTACAACCCGTCCAACAACAAGCGAATCTGAAGAAGCGATGACCACCTCAATCATTACAACCACAATGGACCCTGAAGAAACTATTGTTTTTCCTGAAGACTAG
Protein
MLAGSTFAQSFHNPSLNGEETSTNLGEVESRENAPDLEFQYHDHDQMTRYLRAVSAKYPELTALYSIGKSVQGRDLWVMVVSVSPYEHMIGKPDVKYVANIHGNEAVGRELLLHLIQYLVTSYATDPYVKWLLDNTRIHLMPSMNPDGFAISREGQCDTIHGRHNARRHDLNRNFPDFFKANTKQPQPETEAVKQWISKIQFVLSGSLHGGALVASYPFDNTPSSTVFQNYEHTPSISPDDDVFKHLSLVYSKNHAKMSRGVSCKSGSPRFENGITNGAAWYPLTGGMQDYNYLWHGCMEVTLEISCCKYPPAHELPKYWQDNKQSLIKYLAEVHRGAHGFVMDENNNPVERAAIKIKGRDVNYHTTNFGEFWRILLPGTYRMDISAEGYLPQEVEFIVIDSHPTLLNVTLRSAKDVTTRPTTSESEEAMTTSIITTTMDPEETIVFPED
Summary
Similarity
Belongs to the type-B carboxylesterase/lipase family.
Uniprot
A0A2H1V3E3
A0A194PQK2
A0A212F5K1
A0A194QY63
W8B0P1
A0A1B0CR20
+ More
A0A0K8VPW2 A0A0K8V2T1 A0A0A1XR52 A0A0K8VCR1 A0A034VHI3 A0A0A1WWD8 A0A1B0DJT4 A0A1I8M573 B4M280 A0A182G4V6 B4L1N7 A0A1I8M569 A0A1I8Q574 A0A182GM98 A0A1I8M574 A0A1I8M576 A0A1I8Q580 A0A0Q9WLL3 B4JIV8 A0A0Q9WQ54 A0A0Q9X062 A0A1B1UZG6 B0XKN5 A0A0P6IY98 A0A0Q9WY01 A0A1Q3FNI8 Q9VXC4 A0A0M3QZ87 A0A1I8Q572 B7Z0Z6 A0A1W4VX05 Q16RA6 A0A2H8U1T6 A0A0P8XL69 A0A0L0C9K7 Q29IJ7 A0A023GPN7 B3NVL6 M9NFA0 G4LU56 A0A2S2PCI8 A0A1I8Q562 B4NDT0 A0A1Q3FNE4 A0A1I8Q5A8 B4Q1U0 A0A1S4G7R5 A0A1B0BR12 A0A1W4VIZ4 A0A0P8XL55 A0A182V964 X2JFS7 A0A2J7PJV6 A0A2M4CQM3 B3MWA7 A0A0Q5THM9 A0A182U1Y6 A0A182I9I5 A0A182XEL6 A0A182LRG1 A0A1A9Y2X6 A0A0R1EBE5 A0A1W4VIR3 A0A3B0K5D3 A0A0P9AB63 A0A1B0A9X9 A0A067R6S5 A0A0Q5T3K9 A0A1V1FTI7 W5JLU2 A0A1W4VW68 A0A182Q3Q2 A4V4N7 A0A182XXR0 A0A1B6GXV1 A0A0Q5T3V4 A0A182FIJ0 A0A182RPG6 A0A182NAB7 B7Z0Z5 J9JWQ5 A0A0R1EIA7 A0A2J7PJX9 A0A1B6E9U3 A0A182LIM4 F5HJX9 Q7PRP0 A0A1A9VC85 A0A182W480 A0A224XFS6 A0A2K8JS22 A0A0R1EBL9 A0A182KB53 A0A1B6M867
A0A0K8VPW2 A0A0K8V2T1 A0A0A1XR52 A0A0K8VCR1 A0A034VHI3 A0A0A1WWD8 A0A1B0DJT4 A0A1I8M573 B4M280 A0A182G4V6 B4L1N7 A0A1I8M569 A0A1I8Q574 A0A182GM98 A0A1I8M574 A0A1I8M576 A0A1I8Q580 A0A0Q9WLL3 B4JIV8 A0A0Q9WQ54 A0A0Q9X062 A0A1B1UZG6 B0XKN5 A0A0P6IY98 A0A0Q9WY01 A0A1Q3FNI8 Q9VXC4 A0A0M3QZ87 A0A1I8Q572 B7Z0Z6 A0A1W4VX05 Q16RA6 A0A2H8U1T6 A0A0P8XL69 A0A0L0C9K7 Q29IJ7 A0A023GPN7 B3NVL6 M9NFA0 G4LU56 A0A2S2PCI8 A0A1I8Q562 B4NDT0 A0A1Q3FNE4 A0A1I8Q5A8 B4Q1U0 A0A1S4G7R5 A0A1B0BR12 A0A1W4VIZ4 A0A0P8XL55 A0A182V964 X2JFS7 A0A2J7PJV6 A0A2M4CQM3 B3MWA7 A0A0Q5THM9 A0A182U1Y6 A0A182I9I5 A0A182XEL6 A0A182LRG1 A0A1A9Y2X6 A0A0R1EBE5 A0A1W4VIR3 A0A3B0K5D3 A0A0P9AB63 A0A1B0A9X9 A0A067R6S5 A0A0Q5T3K9 A0A1V1FTI7 W5JLU2 A0A1W4VW68 A0A182Q3Q2 A4V4N7 A0A182XXR0 A0A1B6GXV1 A0A0Q5T3V4 A0A182FIJ0 A0A182RPG6 A0A182NAB7 B7Z0Z5 J9JWQ5 A0A0R1EIA7 A0A2J7PJX9 A0A1B6E9U3 A0A182LIM4 F5HJX9 Q7PRP0 A0A1A9VC85 A0A182W480 A0A224XFS6 A0A2K8JS22 A0A0R1EBL9 A0A182KB53 A0A1B6M867
Pubmed
EMBL
ODYU01000493
SOQ35365.1
KQ459595
KPI95587.1
AGBW02010183
OWR49003.1
+ More
KQ461154 KPJ08516.1 GAMC01011895 JAB94660.1 AJWK01024342 AJWK01024343 GDHF01011431 GDHF01003535 JAI40883.1 JAI48779.1 GDHF01019191 JAI33123.1 GBXI01000523 JAD13769.1 GDHF01015677 JAI36637.1 GAKP01017752 JAC41200.1 GBXI01010933 JAD03359.1 AJVK01001052 AJVK01001053 AJVK01001054 CH940651 EDW65784.2 JXUM01144536 KQ569811 KXJ68512.1 CH933810 EDW07673.2 JXUM01074065 KQ562810 KXJ75079.1 KRF82417.1 CH916370 EDV99522.1 KRF82416.1 KRF94133.1 KX428487 ANW09598.1 DS233924 EDS32354.1 GDUN01000121 JAN95798.1 KRF94132.1 GFDL01005876 JAV29169.1 AE014298 AAF48652.3 CP012528 ALC48949.1 BT132702 ACL82940.1 AEQ72804.2 CH477719 EAT36941.1 GFXV01008047 MBW19852.1 CH902625 KPU75497.1 JRES01000728 KNC28936.1 CH379063 EAL32656.2 ABC67188.2 CH954180 EDV46408.1 AFH07431.1 BT132686 AEQ62990.1 GGMR01014554 MBY27173.1 CH964239 EDW81899.2 GFDL01006042 JAV29003.1 CM000162 EDX02515.1 JXJN01018875 KPU75496.1 AHN59814.1 NEVH01024944 PNF16608.1 GGFL01003401 MBW67579.1 EDV35252.2 KQS29963.1 APCN01000984 AXCM01000900 KRK06812.1 OUUW01000021 SPP89424.1 KPU75495.1 KK852704 KDR18068.1 KQS29964.1 FX985402 BAX07415.1 ADMH02001203 ETN63744.1 AXCN02002441 AAN09418.2 GECZ01002511 JAS67258.1 KQS29965.1 ACL82941.1 ABLF02026859 ABLF02026864 KRK06810.1 PNF16609.1 GEDC01002664 JAS34634.1 AAAB01008847 EGK96595.1 EAA06873.5 GFTR01006542 JAW09884.1 KY031089 ATU82840.1 KRK06811.1 GEBQ01007854 JAT32123.1
KQ461154 KPJ08516.1 GAMC01011895 JAB94660.1 AJWK01024342 AJWK01024343 GDHF01011431 GDHF01003535 JAI40883.1 JAI48779.1 GDHF01019191 JAI33123.1 GBXI01000523 JAD13769.1 GDHF01015677 JAI36637.1 GAKP01017752 JAC41200.1 GBXI01010933 JAD03359.1 AJVK01001052 AJVK01001053 AJVK01001054 CH940651 EDW65784.2 JXUM01144536 KQ569811 KXJ68512.1 CH933810 EDW07673.2 JXUM01074065 KQ562810 KXJ75079.1 KRF82417.1 CH916370 EDV99522.1 KRF82416.1 KRF94133.1 KX428487 ANW09598.1 DS233924 EDS32354.1 GDUN01000121 JAN95798.1 KRF94132.1 GFDL01005876 JAV29169.1 AE014298 AAF48652.3 CP012528 ALC48949.1 BT132702 ACL82940.1 AEQ72804.2 CH477719 EAT36941.1 GFXV01008047 MBW19852.1 CH902625 KPU75497.1 JRES01000728 KNC28936.1 CH379063 EAL32656.2 ABC67188.2 CH954180 EDV46408.1 AFH07431.1 BT132686 AEQ62990.1 GGMR01014554 MBY27173.1 CH964239 EDW81899.2 GFDL01006042 JAV29003.1 CM000162 EDX02515.1 JXJN01018875 KPU75496.1 AHN59814.1 NEVH01024944 PNF16608.1 GGFL01003401 MBW67579.1 EDV35252.2 KQS29963.1 APCN01000984 AXCM01000900 KRK06812.1 OUUW01000021 SPP89424.1 KPU75495.1 KK852704 KDR18068.1 KQS29964.1 FX985402 BAX07415.1 ADMH02001203 ETN63744.1 AXCN02002441 AAN09418.2 GECZ01002511 JAS67258.1 KQS29965.1 ACL82941.1 ABLF02026859 ABLF02026864 KRK06810.1 PNF16609.1 GEDC01002664 JAS34634.1 AAAB01008847 EGK96595.1 EAA06873.5 GFTR01006542 JAW09884.1 KY031089 ATU82840.1 KRK06811.1 GEBQ01007854 JAT32123.1
Proteomes
UP000053268
UP000007151
UP000053240
UP000092461
UP000092462
UP000095301
+ More
UP000008792 UP000069940 UP000249989 UP000009192 UP000095300 UP000001070 UP000002320 UP000000803 UP000092553 UP000192221 UP000008820 UP000007801 UP000037069 UP000001819 UP000008711 UP000007798 UP000002282 UP000092460 UP000075903 UP000235965 UP000075902 UP000075840 UP000076407 UP000075883 UP000092443 UP000268350 UP000092445 UP000027135 UP000000673 UP000075886 UP000076408 UP000069272 UP000075900 UP000075884 UP000007819 UP000075882 UP000007062 UP000078200 UP000075920 UP000075881
UP000008792 UP000069940 UP000249989 UP000009192 UP000095300 UP000001070 UP000002320 UP000000803 UP000092553 UP000192221 UP000008820 UP000007801 UP000037069 UP000001819 UP000008711 UP000007798 UP000002282 UP000092460 UP000075903 UP000235965 UP000075902 UP000075840 UP000076407 UP000075883 UP000092443 UP000268350 UP000092445 UP000027135 UP000000673 UP000075886 UP000076408 UP000069272 UP000075900 UP000075884 UP000007819 UP000075882 UP000007062 UP000078200 UP000075920 UP000075881
PRIDE
Interpro
IPR015567
Pept_M14B_carboxypept_D2
+ More
IPR000834 Peptidase_M14
IPR008969 CarboxyPept-like_regulatory
IPR029058 AB_hydrolase
IPR002018 CarbesteraseB
IPR019826 Carboxylesterase_B_AS
IPR019819 Carboxylesterase_B_CS
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR000664 Lethal2_giant
IPR013577 LLGL2
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR013905 Lgl_C_dom
IPR001388 Synaptobrevin
IPR001680 WD40_repeat
IPR000834 Peptidase_M14
IPR008969 CarboxyPept-like_regulatory
IPR029058 AB_hydrolase
IPR002018 CarbesteraseB
IPR019826 Carboxylesterase_B_AS
IPR019819 Carboxylesterase_B_CS
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR000664 Lethal2_giant
IPR013577 LLGL2
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR013905 Lgl_C_dom
IPR001388 Synaptobrevin
IPR001680 WD40_repeat
Gene 3D
ProteinModelPortal
A0A2H1V3E3
A0A194PQK2
A0A212F5K1
A0A194QY63
W8B0P1
A0A1B0CR20
+ More
A0A0K8VPW2 A0A0K8V2T1 A0A0A1XR52 A0A0K8VCR1 A0A034VHI3 A0A0A1WWD8 A0A1B0DJT4 A0A1I8M573 B4M280 A0A182G4V6 B4L1N7 A0A1I8M569 A0A1I8Q574 A0A182GM98 A0A1I8M574 A0A1I8M576 A0A1I8Q580 A0A0Q9WLL3 B4JIV8 A0A0Q9WQ54 A0A0Q9X062 A0A1B1UZG6 B0XKN5 A0A0P6IY98 A0A0Q9WY01 A0A1Q3FNI8 Q9VXC4 A0A0M3QZ87 A0A1I8Q572 B7Z0Z6 A0A1W4VX05 Q16RA6 A0A2H8U1T6 A0A0P8XL69 A0A0L0C9K7 Q29IJ7 A0A023GPN7 B3NVL6 M9NFA0 G4LU56 A0A2S2PCI8 A0A1I8Q562 B4NDT0 A0A1Q3FNE4 A0A1I8Q5A8 B4Q1U0 A0A1S4G7R5 A0A1B0BR12 A0A1W4VIZ4 A0A0P8XL55 A0A182V964 X2JFS7 A0A2J7PJV6 A0A2M4CQM3 B3MWA7 A0A0Q5THM9 A0A182U1Y6 A0A182I9I5 A0A182XEL6 A0A182LRG1 A0A1A9Y2X6 A0A0R1EBE5 A0A1W4VIR3 A0A3B0K5D3 A0A0P9AB63 A0A1B0A9X9 A0A067R6S5 A0A0Q5T3K9 A0A1V1FTI7 W5JLU2 A0A1W4VW68 A0A182Q3Q2 A4V4N7 A0A182XXR0 A0A1B6GXV1 A0A0Q5T3V4 A0A182FIJ0 A0A182RPG6 A0A182NAB7 B7Z0Z5 J9JWQ5 A0A0R1EIA7 A0A2J7PJX9 A0A1B6E9U3 A0A182LIM4 F5HJX9 Q7PRP0 A0A1A9VC85 A0A182W480 A0A224XFS6 A0A2K8JS22 A0A0R1EBL9 A0A182KB53 A0A1B6M867
A0A0K8VPW2 A0A0K8V2T1 A0A0A1XR52 A0A0K8VCR1 A0A034VHI3 A0A0A1WWD8 A0A1B0DJT4 A0A1I8M573 B4M280 A0A182G4V6 B4L1N7 A0A1I8M569 A0A1I8Q574 A0A182GM98 A0A1I8M574 A0A1I8M576 A0A1I8Q580 A0A0Q9WLL3 B4JIV8 A0A0Q9WQ54 A0A0Q9X062 A0A1B1UZG6 B0XKN5 A0A0P6IY98 A0A0Q9WY01 A0A1Q3FNI8 Q9VXC4 A0A0M3QZ87 A0A1I8Q572 B7Z0Z6 A0A1W4VX05 Q16RA6 A0A2H8U1T6 A0A0P8XL69 A0A0L0C9K7 Q29IJ7 A0A023GPN7 B3NVL6 M9NFA0 G4LU56 A0A2S2PCI8 A0A1I8Q562 B4NDT0 A0A1Q3FNE4 A0A1I8Q5A8 B4Q1U0 A0A1S4G7R5 A0A1B0BR12 A0A1W4VIZ4 A0A0P8XL55 A0A182V964 X2JFS7 A0A2J7PJV6 A0A2M4CQM3 B3MWA7 A0A0Q5THM9 A0A182U1Y6 A0A182I9I5 A0A182XEL6 A0A182LRG1 A0A1A9Y2X6 A0A0R1EBE5 A0A1W4VIR3 A0A3B0K5D3 A0A0P9AB63 A0A1B0A9X9 A0A067R6S5 A0A0Q5T3K9 A0A1V1FTI7 W5JLU2 A0A1W4VW68 A0A182Q3Q2 A4V4N7 A0A182XXR0 A0A1B6GXV1 A0A0Q5T3V4 A0A182FIJ0 A0A182RPG6 A0A182NAB7 B7Z0Z5 J9JWQ5 A0A0R1EIA7 A0A2J7PJX9 A0A1B6E9U3 A0A182LIM4 F5HJX9 Q7PRP0 A0A1A9VC85 A0A182W480 A0A224XFS6 A0A2K8JS22 A0A0R1EBL9 A0A182KB53 A0A1B6M867
PDB
1UWY
E-value=2.0211e-104,
Score=969
Ontologies
GO
PANTHER
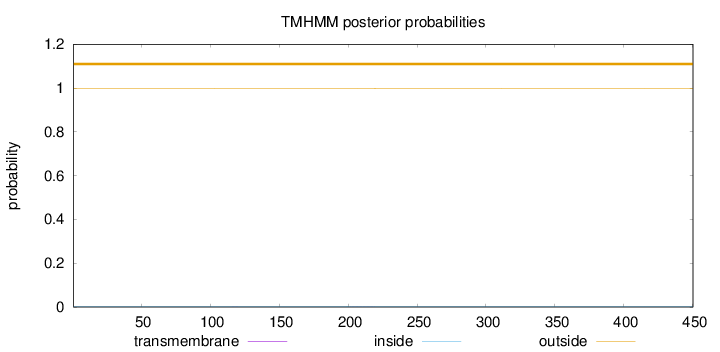
Topology
Length:
450
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00713999999999998
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00189
outside
1 - 450
Population Genetic Test Statistics
Pi
191.129614
Theta
166.654056
Tajima's D
0.876306
CLR
0.322394
CSRT
0.629668516574171
Interpretation
Uncertain
