Pre Gene Modal
BGIBMGA005488
Annotation
replication_factor_C_(activator_1)_5_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.124 Nuclear Reliability : 1.498
Sequence
CDS
ATGACTGTTGGGGAAACAGGCCAAAATTTGCCATGGGTTGAAAAATATCGCCCAAAAAGATTGGATGATTTAGTATCTCATGATGATATAATAAAAACAATAAATCAATTTATGAAAGAGAATCAGCTCCCACATCTACTATTCTACGGTCCTCCTGGAACCGGGAAGACATCTACCATTCTAGCCTGTGCCAAACAAATGTACACTCCACAGCAATTTAGTTCGATGGTGTTGGAACTTAATGCTTCTGACGATAGAGGCATTGGCATTGTTAGAGGTCAGATACTCAGTTTTGCCTCAACAAGGACTATATTTAAAGCAGGACCTAAATTAATTATACTTGATGAAGCTGATGCTATGACAAATGACGCTCAGAATGCACTAAGGAGAATTATAGAGAAGTACACCGAGAATGTAAGGTTTTGCATAATATGCAACTACCTGGGTAAAATTATTCCAGCATTACAGTCGCGGTGTACGCGATTTCGTTTTGCCCCTCTCAAGCAAGATCAAATTGTGCCGAGATTACAAGAAATTGTCACAACGGAAGGTGTAAAAATGTCAGAGGGCGGAATGAAGGCGCTTCTCACCTTGTCCGGGGGTGATATGAGAAAGGTTTTGAATACGCTTCAAAGCACGTGGCTGGCGTATCGTGACGTCACCGAAGATAATGTTTACACTTGCGTCGGCCACCCCCTCCGCGCCGATATCGACAGCATACTCAATTGGTTGCTAAACGAAAATGATTTTTCCGCCTGCTTTAAATCCATACAAGACTTGAAGATTGCAAAAGGATTAGCACTGAGCGATATATTGGCTGAAGTCCATACTATCATACAGAGAGTGAAACTGCCTCCAGAAGTATTAGTGTCATTGCTCATAAAGATGTCAGACGCTGAAGCGAGGTTGGCGAGCGGAAGCAGCGAGCGGGTTGAACTTGCCGCGCTCATCGCGAGCTTCCAGATCGCTAGGGATCAAGTCAAACTAGACACAGAAGTTTCGTAG
Protein
MTVGETGQNLPWVEKYRPKRLDDLVSHDDIIKTINQFMKENQLPHLLFYGPPGTGKTSTILACAKQMYTPQQFSSMVLELNASDDRGIGIVRGQILSFASTRTIFKAGPKLIILDEADAMTNDAQNALRRIIEKYTENVRFCIICNYLGKIIPALQSRCTRFRFAPLKQDQIVPRLQEIVTTEGVKMSEGGMKALLTLSGGDMRKVLNTLQSTWLAYRDVTEDNVYTCVGHPLRADIDSILNWLLNENDFSACFKSIQDLKIAKGLALSDILAEVHTIIQRVKLPPEVLVSLLIKMSDAEARLASGSSERVELAALIASFQIARDQVKLDTEVS
Summary
Uniprot
Q2F659
A0A3S2TNG6
S4P928
A0A2Z5TTY8
A0A1Q3G0M8
A0A067R4E7
+ More
Q16VR5 A0A023ER07 U5ESE9 A0A182HAP6 A0A2J7PJU0 A0A139WCT9 A0A1B6DUV7 A0A2C9JKL7 A0A182N870 A0A1J1IAJ4 V4AWH6 A0A084W2G8 A0A182K9I9 A0A182IMC7 Q7Q9N2 W5JCV6 A0A182LBF5 A0A182HP46 A0A182XJB5 A0A182QQI7 A0A182YNQ6 A0A182V331 A0A182UEZ7 A0A182RTR4 A0A1B6FDY5 K1QQQ5 A0A1W4X2G0 A0A182P5A0 A0A232F3N8 A0A182M4X1 A0A182F2T5 A0A154P2K9 A0A1S3J4Y9 A0A2T7Q049 A0A182WAC9 A0A1B6LAT8 A0A0B7A0Z1 J9K528 A0A0A9YYY3 A0A2A3ELP8 A0A0L7QUI2 A0A088A1X2 F6Y531 A0A2P2I735 Q7ZTM5 Q5XGD2 A0A226PAH3 A0A2H8TJB8 A0A1L8HQV3 A0A2L2Y8Q0 Q6GQ59 A0A3B3S7U3 A0A087USP0 A0A336KMU1 F4WQY4 G3PMC0 H3BBL3 A0A1Y1KP68 A0A3P8XIM5 A0A3Q3VPU7 G3PMB2 A0A3Q3EVH8 A0A1B0AP95 A0A3Q0SRZ7 A0A069DRP0 A0A3B4XMX6 A0A3Q2EBV7 A0A0P5KZI9 A0A0A1WSZ3 A0A1W4Z691 E2AVB8 A0A158NTL4 H3DDU4 Q4RTX1 A0A2U9BU49 A0A034WEI1 A0A3Q2XA33 A0A3Q4HQD6 A0A1B0AK70 V9L2K7 A0A3B4E910 E9HFI1 A0A3B4UZ41 A0A147B151 W8BR34 C3KHF7 R7ULZ2 H9GNG0 Q6DRK4 A0A0P5LST8 A0A0F8CFP6 W5K0V0 A0A3Q3LB57 A0A060Y653
Q16VR5 A0A023ER07 U5ESE9 A0A182HAP6 A0A2J7PJU0 A0A139WCT9 A0A1B6DUV7 A0A2C9JKL7 A0A182N870 A0A1J1IAJ4 V4AWH6 A0A084W2G8 A0A182K9I9 A0A182IMC7 Q7Q9N2 W5JCV6 A0A182LBF5 A0A182HP46 A0A182XJB5 A0A182QQI7 A0A182YNQ6 A0A182V331 A0A182UEZ7 A0A182RTR4 A0A1B6FDY5 K1QQQ5 A0A1W4X2G0 A0A182P5A0 A0A232F3N8 A0A182M4X1 A0A182F2T5 A0A154P2K9 A0A1S3J4Y9 A0A2T7Q049 A0A182WAC9 A0A1B6LAT8 A0A0B7A0Z1 J9K528 A0A0A9YYY3 A0A2A3ELP8 A0A0L7QUI2 A0A088A1X2 F6Y531 A0A2P2I735 Q7ZTM5 Q5XGD2 A0A226PAH3 A0A2H8TJB8 A0A1L8HQV3 A0A2L2Y8Q0 Q6GQ59 A0A3B3S7U3 A0A087USP0 A0A336KMU1 F4WQY4 G3PMC0 H3BBL3 A0A1Y1KP68 A0A3P8XIM5 A0A3Q3VPU7 G3PMB2 A0A3Q3EVH8 A0A1B0AP95 A0A3Q0SRZ7 A0A069DRP0 A0A3B4XMX6 A0A3Q2EBV7 A0A0P5KZI9 A0A0A1WSZ3 A0A1W4Z691 E2AVB8 A0A158NTL4 H3DDU4 Q4RTX1 A0A2U9BU49 A0A034WEI1 A0A3Q2XA33 A0A3Q4HQD6 A0A1B0AK70 V9L2K7 A0A3B4E910 E9HFI1 A0A3B4UZ41 A0A147B151 W8BR34 C3KHF7 R7ULZ2 H9GNG0 Q6DRK4 A0A0P5LST8 A0A0F8CFP6 W5K0V0 A0A3Q3LB57 A0A060Y653
Pubmed
23622113
26760975
24845553
17510324
24945155
26483478
+ More
18362917 19820115 15562597 23254933 24438588 12364791 14747013 17210077 20920257 23761445 20966253 25244985 22992520 28648823 26383154 25401762 26823975 20431018 27762356 24621616 26561354 29240929 21719571 9215903 28004739 25069045 26334808 25830018 20798317 21347285 15496914 25348373 25186727 24402279 21292972 24495485 21881562 15256591 23594743 25835551 25329095 24755649
18362917 19820115 15562597 23254933 24438588 12364791 14747013 17210077 20920257 23761445 20966253 25244985 22992520 28648823 26383154 25401762 26823975 20431018 27762356 24621616 26561354 29240929 21719571 9215903 28004739 25069045 26334808 25830018 20798317 21347285 15496914 25348373 25186727 24402279 21292972 24495485 21881562 15256591 23594743 25835551 25329095 24755649
EMBL
DQ311213
ABD36158.1
RSAL01000041
RVE50849.1
GAIX01005711
JAA86849.1
+ More
FX985787 BBA93674.1 GFDL01001695 JAV33350.1 KK852704 KDR18066.1 CH477583 EAT38657.1 GAPW01002534 JAC11064.1 GANO01002388 JAB57483.1 JXUM01123046 KQ566673 KXJ69915.1 NEVH01024944 PNF16607.1 KQ971362 KYB25740.1 GEDC01007838 JAS29460.1 CVRI01000044 CRK96588.1 KB201304 ESO97856.1 ATLV01019619 KE525275 KFB44412.1 AAAB01008900 EAA09454.3 ADMH02001599 ETN61891.1 APCN01002909 AXCN02001004 GECZ01021378 JAS48391.1 JH816734 EKC39302.1 NNAY01001115 OXU25090.1 AXCM01000427 KQ434792 KZC05458.1 PZQS01000001 PVD39041.1 GEBQ01026776 GEBQ01019236 GEBQ01003757 GEBQ01002845 JAT13201.1 JAT20741.1 JAT36220.1 JAT37132.1 HACG01027708 HACG01027709 CEK74573.1 CEK74574.1 ABLF02026864 GBHO01006190 GBRD01007803 GDHC01013596 JAG37414.1 JAG58018.1 JAQ05033.1 KZ288215 PBC32648.1 KQ414735 KOC62219.1 AAMC01076673 AAMC01076674 AAMC01076675 AAMC01076676 IACF01004241 LAB69835.1 BC044712 CM004466 AAH44712.1 OCU02132.1 BC084510 CR926393 AAH84510.1 CAJ83540.1 AWGT02000122 OXB76459.1 GFXV01002286 MBW14091.1 CM004467 OCT98461.1 IAAA01013400 LAA03710.1 BC072889 AAH72889.1 KK121385 KFM80379.1 UFQS01000530 UFQT01000530 SSX04665.1 SSX25028.1 GL888275 EGI63497.1 AFYH01046357 AFYH01046358 GEZM01082902 JAV61236.1 JXJN01001282 GBGD01002151 JAC86738.1 GDIP01169486 GDIQ01185072 LRGB01000115 JAJ53916.1 JAK66653.1 KZS20756.1 GBXI01012667 JAD01625.1 GL443064 EFN62614.1 ADTU01026005 CAAE01014996 CAG08161.1 CP026251 AWP07571.1 GAKP01006774 JAC52178.1 JW873112 AFP05630.1 GL732636 EFX69502.1 GCES01002271 JAR84052.1 GAMC01002800 JAC03756.1 BT082372 ACQ58079.1 AMQN01007197 KB300180 ELU07083.1 CU207223 AY648755 AAT68073.1 GDIQ01168829 JAK82896.1 KQ042181 KKF18331.1 FR907743 CDQ87206.1
FX985787 BBA93674.1 GFDL01001695 JAV33350.1 KK852704 KDR18066.1 CH477583 EAT38657.1 GAPW01002534 JAC11064.1 GANO01002388 JAB57483.1 JXUM01123046 KQ566673 KXJ69915.1 NEVH01024944 PNF16607.1 KQ971362 KYB25740.1 GEDC01007838 JAS29460.1 CVRI01000044 CRK96588.1 KB201304 ESO97856.1 ATLV01019619 KE525275 KFB44412.1 AAAB01008900 EAA09454.3 ADMH02001599 ETN61891.1 APCN01002909 AXCN02001004 GECZ01021378 JAS48391.1 JH816734 EKC39302.1 NNAY01001115 OXU25090.1 AXCM01000427 KQ434792 KZC05458.1 PZQS01000001 PVD39041.1 GEBQ01026776 GEBQ01019236 GEBQ01003757 GEBQ01002845 JAT13201.1 JAT20741.1 JAT36220.1 JAT37132.1 HACG01027708 HACG01027709 CEK74573.1 CEK74574.1 ABLF02026864 GBHO01006190 GBRD01007803 GDHC01013596 JAG37414.1 JAG58018.1 JAQ05033.1 KZ288215 PBC32648.1 KQ414735 KOC62219.1 AAMC01076673 AAMC01076674 AAMC01076675 AAMC01076676 IACF01004241 LAB69835.1 BC044712 CM004466 AAH44712.1 OCU02132.1 BC084510 CR926393 AAH84510.1 CAJ83540.1 AWGT02000122 OXB76459.1 GFXV01002286 MBW14091.1 CM004467 OCT98461.1 IAAA01013400 LAA03710.1 BC072889 AAH72889.1 KK121385 KFM80379.1 UFQS01000530 UFQT01000530 SSX04665.1 SSX25028.1 GL888275 EGI63497.1 AFYH01046357 AFYH01046358 GEZM01082902 JAV61236.1 JXJN01001282 GBGD01002151 JAC86738.1 GDIP01169486 GDIQ01185072 LRGB01000115 JAJ53916.1 JAK66653.1 KZS20756.1 GBXI01012667 JAD01625.1 GL443064 EFN62614.1 ADTU01026005 CAAE01014996 CAG08161.1 CP026251 AWP07571.1 GAKP01006774 JAC52178.1 JW873112 AFP05630.1 GL732636 EFX69502.1 GCES01002271 JAR84052.1 GAMC01002800 JAC03756.1 BT082372 ACQ58079.1 AMQN01007197 KB300180 ELU07083.1 CU207223 AY648755 AAT68073.1 GDIQ01168829 JAK82896.1 KQ042181 KKF18331.1 FR907743 CDQ87206.1
Proteomes
UP000283053
UP000027135
UP000008820
UP000069940
UP000249989
UP000235965
+ More
UP000007266 UP000076420 UP000075884 UP000183832 UP000030746 UP000030765 UP000075881 UP000075880 UP000007062 UP000000673 UP000075882 UP000075840 UP000076407 UP000075886 UP000076408 UP000075903 UP000075902 UP000075900 UP000005408 UP000192223 UP000075885 UP000215335 UP000075883 UP000069272 UP000076502 UP000085678 UP000245119 UP000075920 UP000007819 UP000242457 UP000053825 UP000005203 UP000008143 UP000186698 UP000198419 UP000261540 UP000054359 UP000007755 UP000007635 UP000008672 UP000265140 UP000261620 UP000261660 UP000092460 UP000261340 UP000261360 UP000265020 UP000076858 UP000192224 UP000000311 UP000005205 UP000007303 UP000246464 UP000264820 UP000261580 UP000092445 UP000261440 UP000000305 UP000261420 UP000265000 UP000014760 UP000001646 UP000000437 UP000018467 UP000261640 UP000193380
UP000007266 UP000076420 UP000075884 UP000183832 UP000030746 UP000030765 UP000075881 UP000075880 UP000007062 UP000000673 UP000075882 UP000075840 UP000076407 UP000075886 UP000076408 UP000075903 UP000075902 UP000075900 UP000005408 UP000192223 UP000075885 UP000215335 UP000075883 UP000069272 UP000076502 UP000085678 UP000245119 UP000075920 UP000007819 UP000242457 UP000053825 UP000005203 UP000008143 UP000186698 UP000198419 UP000261540 UP000054359 UP000007755 UP000007635 UP000008672 UP000265140 UP000261620 UP000261660 UP000092460 UP000261340 UP000261360 UP000265020 UP000076858 UP000192224 UP000000311 UP000005205 UP000007303 UP000246464 UP000264820 UP000261580 UP000092445 UP000261440 UP000000305 UP000261420 UP000265000 UP000014760 UP000001646 UP000000437 UP000018467 UP000261640 UP000193380
PRIDE
Interpro
ProteinModelPortal
Q2F659
A0A3S2TNG6
S4P928
A0A2Z5TTY8
A0A1Q3G0M8
A0A067R4E7
+ More
Q16VR5 A0A023ER07 U5ESE9 A0A182HAP6 A0A2J7PJU0 A0A139WCT9 A0A1B6DUV7 A0A2C9JKL7 A0A182N870 A0A1J1IAJ4 V4AWH6 A0A084W2G8 A0A182K9I9 A0A182IMC7 Q7Q9N2 W5JCV6 A0A182LBF5 A0A182HP46 A0A182XJB5 A0A182QQI7 A0A182YNQ6 A0A182V331 A0A182UEZ7 A0A182RTR4 A0A1B6FDY5 K1QQQ5 A0A1W4X2G0 A0A182P5A0 A0A232F3N8 A0A182M4X1 A0A182F2T5 A0A154P2K9 A0A1S3J4Y9 A0A2T7Q049 A0A182WAC9 A0A1B6LAT8 A0A0B7A0Z1 J9K528 A0A0A9YYY3 A0A2A3ELP8 A0A0L7QUI2 A0A088A1X2 F6Y531 A0A2P2I735 Q7ZTM5 Q5XGD2 A0A226PAH3 A0A2H8TJB8 A0A1L8HQV3 A0A2L2Y8Q0 Q6GQ59 A0A3B3S7U3 A0A087USP0 A0A336KMU1 F4WQY4 G3PMC0 H3BBL3 A0A1Y1KP68 A0A3P8XIM5 A0A3Q3VPU7 G3PMB2 A0A3Q3EVH8 A0A1B0AP95 A0A3Q0SRZ7 A0A069DRP0 A0A3B4XMX6 A0A3Q2EBV7 A0A0P5KZI9 A0A0A1WSZ3 A0A1W4Z691 E2AVB8 A0A158NTL4 H3DDU4 Q4RTX1 A0A2U9BU49 A0A034WEI1 A0A3Q2XA33 A0A3Q4HQD6 A0A1B0AK70 V9L2K7 A0A3B4E910 E9HFI1 A0A3B4UZ41 A0A147B151 W8BR34 C3KHF7 R7ULZ2 H9GNG0 Q6DRK4 A0A0P5LST8 A0A0F8CFP6 W5K0V0 A0A3Q3LB57 A0A060Y653
Q16VR5 A0A023ER07 U5ESE9 A0A182HAP6 A0A2J7PJU0 A0A139WCT9 A0A1B6DUV7 A0A2C9JKL7 A0A182N870 A0A1J1IAJ4 V4AWH6 A0A084W2G8 A0A182K9I9 A0A182IMC7 Q7Q9N2 W5JCV6 A0A182LBF5 A0A182HP46 A0A182XJB5 A0A182QQI7 A0A182YNQ6 A0A182V331 A0A182UEZ7 A0A182RTR4 A0A1B6FDY5 K1QQQ5 A0A1W4X2G0 A0A182P5A0 A0A232F3N8 A0A182M4X1 A0A182F2T5 A0A154P2K9 A0A1S3J4Y9 A0A2T7Q049 A0A182WAC9 A0A1B6LAT8 A0A0B7A0Z1 J9K528 A0A0A9YYY3 A0A2A3ELP8 A0A0L7QUI2 A0A088A1X2 F6Y531 A0A2P2I735 Q7ZTM5 Q5XGD2 A0A226PAH3 A0A2H8TJB8 A0A1L8HQV3 A0A2L2Y8Q0 Q6GQ59 A0A3B3S7U3 A0A087USP0 A0A336KMU1 F4WQY4 G3PMC0 H3BBL3 A0A1Y1KP68 A0A3P8XIM5 A0A3Q3VPU7 G3PMB2 A0A3Q3EVH8 A0A1B0AP95 A0A3Q0SRZ7 A0A069DRP0 A0A3B4XMX6 A0A3Q2EBV7 A0A0P5KZI9 A0A0A1WSZ3 A0A1W4Z691 E2AVB8 A0A158NTL4 H3DDU4 Q4RTX1 A0A2U9BU49 A0A034WEI1 A0A3Q2XA33 A0A3Q4HQD6 A0A1B0AK70 V9L2K7 A0A3B4E910 E9HFI1 A0A3B4UZ41 A0A147B151 W8BR34 C3KHF7 R7ULZ2 H9GNG0 Q6DRK4 A0A0P5LST8 A0A0F8CFP6 W5K0V0 A0A3Q3LB57 A0A060Y653
PDB
1SXJ
E-value=5.75804e-84,
Score=791
Ontologies
PATHWAY
GO
Topology
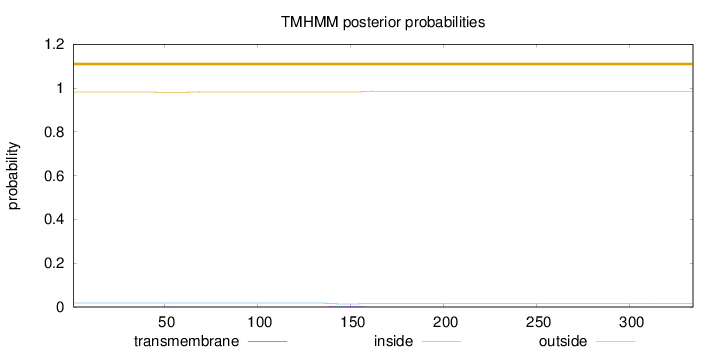
Length:
334
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0898399999999999
Exp number, first 60 AAs:
0.00854
Total prob of N-in:
0.01853
outside
1 - 334
Population Genetic Test Statistics
Pi
256.859622
Theta
186.202382
Tajima's D
1.235235
CLR
0
CSRT
0.720813959302035
Interpretation
Uncertain
