Gene
KWMTBOMO04594 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005323
Annotation
ALY_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.242
Sequence
CDS
ATGGTTGATCAAATTGAAATGGCACTGGATGATATTATAAAAGCCAACAAAAAGACAAGGATTGGAGGTGGTGCTGGAAGAAAATTTGATGCAAACAAAAGAACAGGACGTGGAGGCGGTGCTGGATTTCGTAGCAACCGCTCTGGTGGTGTACAACGAGGTAGAAATCGTGGAGGCATAACGAAATCTACCAACTATTCGAGGGGTGATGTGAACAGCACTTGGAAACATGATATGTTCAATGATTTTGGTGAGAGGAAACTTCAAAGAAACAGTGGGACCATAACAACTGGTCCTACCAAGCTGCTTGTATCCAATTTGGACTTTGGAGTATCAGATTCAGATATAAAGGAACTATTTTCGGAATTTGGCATCCTCAAAAGTGCATCTGTACATTATGATCGGTCAGGTAGATCTTTAGGTACTGCAGATGTTGTGTTTGAAAGAAAAGCAGATGCTGTGAAGGCTATGAAACAGTATAACGGAGTGCCACTTGATGGACGGGCAATGAATATACAATTGGCAACATCTGAAATCAATACTTTAAGGCCCGAGGCTAGAAATCGAATTGGTGGTGCGAGTGTGGGAGGTACTATACGAAACAATCCTAAAAGAGGAGGTGGAGGAATTAGAAATCAAAGCAATAGTGGAGCACGTGGGGGAAACAGAAGAGGACGTGGTGGTGCAGGCAGAGGCAAACGACCAGTACCTACTGCAGAACAATTAGATGCAGAGCTTGATGCATATGTGAAAGAAATCAAGTGA
Protein
MVDQIEMALDDIIKANKKTRIGGGAGRKFDANKRTGRGGGAGFRSNRSGGVQRGRNRGGITKSTNYSRGDVNSTWKHDMFNDFGERKLQRNSGTITTGPTKLLVSNLDFGVSDSDIKELFSEFGILKSASVHYDRSGRSLGTADVVFERKADAVKAMKQYNGVPLDGRAMNIQLATSEINTLRPEARNRIGGASVGGTIRNNPKRGGGGIRNQSNSGARGGNRRGRGGAGRGKRPVPTAEQLDAELDAYVKEIK
Summary
Uniprot
Q1HE02
E1AW12
A0A2A4IVF5
A0A194QSL2
A0A194PRK5
A0A2H1X3F8
+ More
A0A212FPS4 A0A0L7LNF9 A0A3S2NLP4 A0A154P6L7 A0A2A3ESD0 S4PXR6 A0A1L8DHB6 A0A088AH65 A0A0N1ITJ0 A0A1B0CTC0 A0A146LHH0 A0A0L7R9G8 A0A1Y1MVQ3 N6UAL5 V5I8V5 A0A026WLL2 A0A2P8Z9S7 E2ADL0 A0A1W4XE25 A0A1B0DNA7 A0A067QLD4 A0A195E5X6 E9IQT3 A0A195BA74 E2BUS2 A0A195EUU0 F4X7Q4 A0A195D0H6 A0A151X6D4 A0A1Q3EXQ0 R4UJ53 A0A023ELN1 A0A2J7RS03 A0A1B0C3L7 A0A1A9Z9V0 A0A0K8TM95 D3TME7 A0A0T6B4N7 A0A1A9VL32 U4U061 A0A1L8DG42 W5JRY2 B4PS22 B3P2S4 A0A182MUD5 U5ELI9 Q9V3E7 A0A182X204 A0A182L488 A0A182I667 A0A182S020 A0A336LWX8 A0A182WIA1 Q16ED0 B4I4N3 Q7QI92 A0A182UY82 B4QYZ5 B0WLB4 A0A182FI40 A0A1A9WLV3 A0A182N7N0 A0A023F2E4 D6X2T6 A0A182QLX3 A0A1I8NU24 A0A069DQ92 A0A2M4BY31 A0A182PHV2 A0A0P4VL88 T1HS23 A0A1W4UFF4 A0A2M4ABE5 A0A182JXN5 A0A1L8EDI4 A0A158NPJ2 A0A1B6GMN9 A0A224XMG4 A0A0K8UBD0 A0A034WRF0 A0A1W4USK3 A0A182YNL2 A0A1B6GXN9 A0A1J1HS57 A0A182TDU0 B4LZC6 T1PBJ0 A0A1I8NDB1 A0A0A9ZI31 A0A146L8V9 Q293K7 B4GM00 B3LYC2
A0A212FPS4 A0A0L7LNF9 A0A3S2NLP4 A0A154P6L7 A0A2A3ESD0 S4PXR6 A0A1L8DHB6 A0A088AH65 A0A0N1ITJ0 A0A1B0CTC0 A0A146LHH0 A0A0L7R9G8 A0A1Y1MVQ3 N6UAL5 V5I8V5 A0A026WLL2 A0A2P8Z9S7 E2ADL0 A0A1W4XE25 A0A1B0DNA7 A0A067QLD4 A0A195E5X6 E9IQT3 A0A195BA74 E2BUS2 A0A195EUU0 F4X7Q4 A0A195D0H6 A0A151X6D4 A0A1Q3EXQ0 R4UJ53 A0A023ELN1 A0A2J7RS03 A0A1B0C3L7 A0A1A9Z9V0 A0A0K8TM95 D3TME7 A0A0T6B4N7 A0A1A9VL32 U4U061 A0A1L8DG42 W5JRY2 B4PS22 B3P2S4 A0A182MUD5 U5ELI9 Q9V3E7 A0A182X204 A0A182L488 A0A182I667 A0A182S020 A0A336LWX8 A0A182WIA1 Q16ED0 B4I4N3 Q7QI92 A0A182UY82 B4QYZ5 B0WLB4 A0A182FI40 A0A1A9WLV3 A0A182N7N0 A0A023F2E4 D6X2T6 A0A182QLX3 A0A1I8NU24 A0A069DQ92 A0A2M4BY31 A0A182PHV2 A0A0P4VL88 T1HS23 A0A1W4UFF4 A0A2M4ABE5 A0A182JXN5 A0A1L8EDI4 A0A158NPJ2 A0A1B6GMN9 A0A224XMG4 A0A0K8UBD0 A0A034WRF0 A0A1W4USK3 A0A182YNL2 A0A1B6GXN9 A0A1J1HS57 A0A182TDU0 B4LZC6 T1PBJ0 A0A1I8NDB1 A0A0A9ZI31 A0A146L8V9 Q293K7 B4GM00 B3LYC2
Pubmed
19121390
26354079
22118469
26227816
23622113
26823975
+ More
28004739 23537049 24508170 30249741 29403074 20798317 24845553 21282665 21719571 24945155 26483478 26369729 20353571 20920257 23761445 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20966253 17510324 12364791 14747013 17210077 25474469 18362917 19820115 26334808 27129103 21347285 25348373 25244985 25315136 25401762 15632085
28004739 23537049 24508170 30249741 29403074 20798317 24845553 21282665 21719571 24945155 26483478 26369729 20353571 20920257 23761445 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20966253 17510324 12364791 14747013 17210077 25474469 18362917 19820115 26334808 27129103 21347285 25348373 25244985 25315136 25401762 15632085
EMBL
BABH01013933
DQ497195
ABF55960.1
HQ005513
ADM33940.1
NWSH01005989
+ More
PCG63765.1 KQ461154 KPJ08513.1 KQ459595 KPI95583.1 ODYU01013104 SOQ59788.1 AGBW02000920 OWR55737.1 JTDY01000457 KOB77078.1 RSAL01000041 RVE50847.1 KQ434827 KZC07569.1 KZ288189 PBC34665.1 GAIX01004573 JAA87987.1 GFDF01008235 JAV05849.1 KQ435794 KOX73851.1 AJWK01027427 GDHC01012247 JAQ06382.1 KQ414626 KOC67520.1 GEZM01022998 JAV88620.1 APGK01042496 APGK01042497 KB741006 ENN75662.1 GALX01004025 JAB64441.1 KK107182 QOIP01000003 EZA55989.1 RLU24573.1 PYGN01000134 PSN53253.1 GL438820 EFN68372.1 AJVK01007475 AJVK01007476 AJVK01007477 KK853292 KDR08846.1 KQ979608 KYN20287.1 GL764900 EFZ17092.1 KQ976542 KYM81132.1 GL450746 EFN80545.1 KQ981965 KYN31662.1 GL888873 EGI57527.1 KQ977004 KYN06420.1 KQ982482 KYQ55838.1 GFDL01014999 JAV20046.1 KC632260 AGM32074.1 JXUM01009515 JXUM01009516 GAPW01003446 KQ560285 JAC10152.1 KXJ83282.1 NEVH01000596 PNF43614.1 JXJN01025026 GDAI01002116 JAI15487.1 CCAG010002358 EZ422599 ADD18875.1 LJIG01009806 KRT82344.1 KB630127 ERL83420.1 GFDF01008727 JAV05357.1 ADMH02000632 ETN65494.1 CM000160 EDW96422.1 CH954181 EDV48096.1 AXCM01001601 GANO01001385 JAB58486.1 AF172637 AE014297 AAD46930.1 AAF54070.1 APCN01003569 UFQS01000185 UFQT01000185 SSX00889.1 SSX21269.1 CH478842 CH477327 EAT32586.1 EAT43446.1 CH480821 EDW55176.1 AAAB01008807 EAA04421.3 CM000364 EDX11935.1 DS231983 EDS30398.1 GBBI01003528 JAC15184.1 KQ971372 EFA09827.1 AXCN02000581 GBGD01002656 JAC86233.1 GGFJ01008828 MBW57969.1 GDKW01003228 JAI53367.1 ACPB03009184 GGFK01004758 MBW38079.1 GFDG01002028 JAV16771.1 ADTU01022425 ADTU01022426 ADTU01022427 GECZ01013353 GECZ01006061 GECZ01002426 JAS56416.1 JAS63708.1 JAS67343.1 GFTR01004152 JAW12274.1 GDHF01028365 JAI23949.1 GAKP01002197 JAC56755.1 GECZ01002580 JAS67189.1 CVRI01000020 CRK90853.1 CH940650 EDW68161.1 KA646034 AFP60663.1 GBHO01001259 JAG42345.1 GDHC01015529 JAQ03100.1 CM000070 EAL29208.2 CH479185 EDW38574.1 CH902617 EDV41785.1
PCG63765.1 KQ461154 KPJ08513.1 KQ459595 KPI95583.1 ODYU01013104 SOQ59788.1 AGBW02000920 OWR55737.1 JTDY01000457 KOB77078.1 RSAL01000041 RVE50847.1 KQ434827 KZC07569.1 KZ288189 PBC34665.1 GAIX01004573 JAA87987.1 GFDF01008235 JAV05849.1 KQ435794 KOX73851.1 AJWK01027427 GDHC01012247 JAQ06382.1 KQ414626 KOC67520.1 GEZM01022998 JAV88620.1 APGK01042496 APGK01042497 KB741006 ENN75662.1 GALX01004025 JAB64441.1 KK107182 QOIP01000003 EZA55989.1 RLU24573.1 PYGN01000134 PSN53253.1 GL438820 EFN68372.1 AJVK01007475 AJVK01007476 AJVK01007477 KK853292 KDR08846.1 KQ979608 KYN20287.1 GL764900 EFZ17092.1 KQ976542 KYM81132.1 GL450746 EFN80545.1 KQ981965 KYN31662.1 GL888873 EGI57527.1 KQ977004 KYN06420.1 KQ982482 KYQ55838.1 GFDL01014999 JAV20046.1 KC632260 AGM32074.1 JXUM01009515 JXUM01009516 GAPW01003446 KQ560285 JAC10152.1 KXJ83282.1 NEVH01000596 PNF43614.1 JXJN01025026 GDAI01002116 JAI15487.1 CCAG010002358 EZ422599 ADD18875.1 LJIG01009806 KRT82344.1 KB630127 ERL83420.1 GFDF01008727 JAV05357.1 ADMH02000632 ETN65494.1 CM000160 EDW96422.1 CH954181 EDV48096.1 AXCM01001601 GANO01001385 JAB58486.1 AF172637 AE014297 AAD46930.1 AAF54070.1 APCN01003569 UFQS01000185 UFQT01000185 SSX00889.1 SSX21269.1 CH478842 CH477327 EAT32586.1 EAT43446.1 CH480821 EDW55176.1 AAAB01008807 EAA04421.3 CM000364 EDX11935.1 DS231983 EDS30398.1 GBBI01003528 JAC15184.1 KQ971372 EFA09827.1 AXCN02000581 GBGD01002656 JAC86233.1 GGFJ01008828 MBW57969.1 GDKW01003228 JAI53367.1 ACPB03009184 GGFK01004758 MBW38079.1 GFDG01002028 JAV16771.1 ADTU01022425 ADTU01022426 ADTU01022427 GECZ01013353 GECZ01006061 GECZ01002426 JAS56416.1 JAS63708.1 JAS67343.1 GFTR01004152 JAW12274.1 GDHF01028365 JAI23949.1 GAKP01002197 JAC56755.1 GECZ01002580 JAS67189.1 CVRI01000020 CRK90853.1 CH940650 EDW68161.1 KA646034 AFP60663.1 GBHO01001259 JAG42345.1 GDHC01015529 JAQ03100.1 CM000070 EAL29208.2 CH479185 EDW38574.1 CH902617 EDV41785.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000283053 UP000076502 UP000242457 UP000005203 UP000053105 UP000092461 UP000053825 UP000019118 UP000053097 UP000279307 UP000245037 UP000000311 UP000192223 UP000092462 UP000027135 UP000078492 UP000078540 UP000008237 UP000078541 UP000007755 UP000078542 UP000075809 UP000069940 UP000249989 UP000235965 UP000092460 UP000092445 UP000092444 UP000078200 UP000030742 UP000000673 UP000002282 UP000008711 UP000075883 UP000000803 UP000076407 UP000075882 UP000075840 UP000075900 UP000075920 UP000008820 UP000001292 UP000007062 UP000075903 UP000000304 UP000002320 UP000069272 UP000091820 UP000075884 UP000007266 UP000075886 UP000095300 UP000075885 UP000015103 UP000192221 UP000075881 UP000005205 UP000076408 UP000183832 UP000075902 UP000008792 UP000095301 UP000001819 UP000008744 UP000007801
UP000283053 UP000076502 UP000242457 UP000005203 UP000053105 UP000092461 UP000053825 UP000019118 UP000053097 UP000279307 UP000245037 UP000000311 UP000192223 UP000092462 UP000027135 UP000078492 UP000078540 UP000008237 UP000078541 UP000007755 UP000078542 UP000075809 UP000069940 UP000249989 UP000235965 UP000092460 UP000092445 UP000092444 UP000078200 UP000030742 UP000000673 UP000002282 UP000008711 UP000075883 UP000000803 UP000076407 UP000075882 UP000075840 UP000075900 UP000075920 UP000008820 UP000001292 UP000007062 UP000075903 UP000000304 UP000002320 UP000069272 UP000091820 UP000075884 UP000007266 UP000075886 UP000095300 UP000075885 UP000015103 UP000192221 UP000075881 UP000005205 UP000076408 UP000183832 UP000075902 UP000008792 UP000095301 UP000001819 UP000008744 UP000007801
Interpro
SUPFAM
SSF54928
SSF54928
Gene 3D
ProteinModelPortal
Q1HE02
E1AW12
A0A2A4IVF5
A0A194QSL2
A0A194PRK5
A0A2H1X3F8
+ More
A0A212FPS4 A0A0L7LNF9 A0A3S2NLP4 A0A154P6L7 A0A2A3ESD0 S4PXR6 A0A1L8DHB6 A0A088AH65 A0A0N1ITJ0 A0A1B0CTC0 A0A146LHH0 A0A0L7R9G8 A0A1Y1MVQ3 N6UAL5 V5I8V5 A0A026WLL2 A0A2P8Z9S7 E2ADL0 A0A1W4XE25 A0A1B0DNA7 A0A067QLD4 A0A195E5X6 E9IQT3 A0A195BA74 E2BUS2 A0A195EUU0 F4X7Q4 A0A195D0H6 A0A151X6D4 A0A1Q3EXQ0 R4UJ53 A0A023ELN1 A0A2J7RS03 A0A1B0C3L7 A0A1A9Z9V0 A0A0K8TM95 D3TME7 A0A0T6B4N7 A0A1A9VL32 U4U061 A0A1L8DG42 W5JRY2 B4PS22 B3P2S4 A0A182MUD5 U5ELI9 Q9V3E7 A0A182X204 A0A182L488 A0A182I667 A0A182S020 A0A336LWX8 A0A182WIA1 Q16ED0 B4I4N3 Q7QI92 A0A182UY82 B4QYZ5 B0WLB4 A0A182FI40 A0A1A9WLV3 A0A182N7N0 A0A023F2E4 D6X2T6 A0A182QLX3 A0A1I8NU24 A0A069DQ92 A0A2M4BY31 A0A182PHV2 A0A0P4VL88 T1HS23 A0A1W4UFF4 A0A2M4ABE5 A0A182JXN5 A0A1L8EDI4 A0A158NPJ2 A0A1B6GMN9 A0A224XMG4 A0A0K8UBD0 A0A034WRF0 A0A1W4USK3 A0A182YNL2 A0A1B6GXN9 A0A1J1HS57 A0A182TDU0 B4LZC6 T1PBJ0 A0A1I8NDB1 A0A0A9ZI31 A0A146L8V9 Q293K7 B4GM00 B3LYC2
A0A212FPS4 A0A0L7LNF9 A0A3S2NLP4 A0A154P6L7 A0A2A3ESD0 S4PXR6 A0A1L8DHB6 A0A088AH65 A0A0N1ITJ0 A0A1B0CTC0 A0A146LHH0 A0A0L7R9G8 A0A1Y1MVQ3 N6UAL5 V5I8V5 A0A026WLL2 A0A2P8Z9S7 E2ADL0 A0A1W4XE25 A0A1B0DNA7 A0A067QLD4 A0A195E5X6 E9IQT3 A0A195BA74 E2BUS2 A0A195EUU0 F4X7Q4 A0A195D0H6 A0A151X6D4 A0A1Q3EXQ0 R4UJ53 A0A023ELN1 A0A2J7RS03 A0A1B0C3L7 A0A1A9Z9V0 A0A0K8TM95 D3TME7 A0A0T6B4N7 A0A1A9VL32 U4U061 A0A1L8DG42 W5JRY2 B4PS22 B3P2S4 A0A182MUD5 U5ELI9 Q9V3E7 A0A182X204 A0A182L488 A0A182I667 A0A182S020 A0A336LWX8 A0A182WIA1 Q16ED0 B4I4N3 Q7QI92 A0A182UY82 B4QYZ5 B0WLB4 A0A182FI40 A0A1A9WLV3 A0A182N7N0 A0A023F2E4 D6X2T6 A0A182QLX3 A0A1I8NU24 A0A069DQ92 A0A2M4BY31 A0A182PHV2 A0A0P4VL88 T1HS23 A0A1W4UFF4 A0A2M4ABE5 A0A182JXN5 A0A1L8EDI4 A0A158NPJ2 A0A1B6GMN9 A0A224XMG4 A0A0K8UBD0 A0A034WRF0 A0A1W4USK3 A0A182YNL2 A0A1B6GXN9 A0A1J1HS57 A0A182TDU0 B4LZC6 T1PBJ0 A0A1I8NDB1 A0A0A9ZI31 A0A146L8V9 Q293K7 B4GM00 B3LYC2
PDB
2F3J
E-value=1.08593e-33,
Score=356
Ontologies
PATHWAY
GO
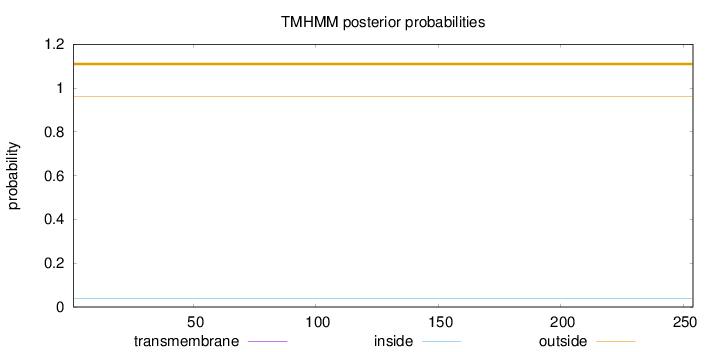
Topology
Length:
254
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03802
outside
1 - 254
Population Genetic Test Statistics
Pi
188.070768
Theta
173.527734
Tajima's D
0.120254
CLR
0.622527
CSRT
0.403929803509824
Interpretation
Uncertain
