Gene
KWMTBOMO04593
Pre Gene Modal
BGIBMGA005324
Annotation
hypothetical_protein_KGM_20770_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 3.54
Sequence
CDS
ATGAACCTCTCAGATACATCAGATGAAGGTTCCAGCAGTGAAGAAAGCAGAAAACCAGCGAAGTGCTCGAGAAGCAATACATATGGTGCATCTTCAACTACATCACATGGATCTGCAATACAATCAGAAGATGAACCTGATACAGCAAATGAATATCATTCAAACCTCAGTACAAAACGAAGAAAAACCGAAATAGAAAATAGCACAGATCGAAATGTGAGATTAAGCAGTAGAAGCAGTATAAGTGAAAACTGCTCGTCGCCCGGCGAACACTTTGAAGATTACCGTCCTGTATTTAATGACAACGATGAAGACCAAGAGGAGAACGCATTACGACCAAGCTTTCAAAATTCTGATACAAAATTTAACAAATCTGTGAATGACATGAATGATCCCGAATATGCTTATACTGGAGAATATTCAGAAAAGTCAAAGCGAATGATGAGTAATATGGGTTACAAGCCGGGTAAAGGTCTTGGTAAATTTGAACATGGACGAACAGAGCCCGTTGAAGCTTCTGCACAAAAAGGCCGAAGAGGATTGGGCTTAAAACCTTCTATTGTTGGTAGTGTACCTGAGGACTTTAAATGGACTCCAGATGAATCCAAACCAGAGGCAAAAGAAAAAGTTATCTGGATGAGTCCCGCCCCAGATGAACCGCTTTCTGGTGACCAGTTGGAGGAATGGTTGAAGAAGGGATCAAAAAAGTTGACTATAGATGACGAAACAGACTTTGTAGAACCCCATATTCTAAAAGGAGTTTTGAAAGCAAAGACAATCTTTGATGATTTAGATGACGAAGACCTGAGGCATGCTCGTTTTAGAAGCAATCCATATGAAACTATTGGTTCAGTGATATTTTTGAACCGGGCTGCCGTTAAAATGGCGAACATAGATGCAGTTTGTGATTTTGTCTTCACTAATCCAAAAAAACAAAATGGAGAAGAAGTCATAGACAAAAAAGAGATCTTGTATTTTGCTGATGTATGTGCTGGTCCTGGTGGCTTTTCAGAATACATTCTATATCGTAGAGGGTGGAGAGCAAAAGGATTTGGTTTTACTCTAAAAGGACCCAACGATTTTAAGCTATCTGACTTTTACGCTGGACCACCAGAAACATTCCACCCCTATTATGGTACGAAAAACGACGGTAACATATTCGATCCATCAAATCTAATGTCACTAAAGGAGCACGTTTTGCAACAAACTGAAGACAAAGGCGTACATTTTTTAATGGCCGATGGGGGATTTTCAGTAGAGGGACAGGAAAACATTCAAGAAATACTTTCGAAGCAGTTGTATTTGTGTCAATGCCTCGCGGCACTGATGCTCGTGAGAACCGGCGGCCATTTTGTGGTGAAACTATTTGATGTCTTCACACGGTTTAGCATTGGCTTGATCTACATTATGTACAGATGTTTTGATAAAGTTTGTATTCATAAACCAGTGACTTCCAGACCTGCAAATTCTGAGAGGTACCTGATTTGTCTGTGGAAGAGGTCATGGACCGAGTCAGCAGAACATCACCTTTACACTGTGAACTCGATCCTATGGAACAGTACTAACAGTGATATTGATATTTTAGAACTAGTTCCTATTGACATCATACAGGAAGATACTGAATTTTTCGAATACGTTTACAAGAGTAACTGCATTCTCGGCGAAAGGCAAATACAAAATTTACTTAAGATAGCTGCATTTAGCAAACACAAAATTTTGAAAGAAATTTATCAAGCTGAAATGAAAGAACAGTGCTTGCAACTCTGGAATTTGCCAGATCAAGTCAGAACTCAACCAAAACGCCTCAATCCCGATGAAACATTAAAAAACATTTTGTCCACATTAAAAACACCCAATCCTCACAGTCTGTTCAATTTCAAAACGAATATATTAAACAAGAAACCTGAATACATAGAGGTGGAAGGTGATTTGACAAATTTCAAGAATGTATACGACTGGCACTTCATGTTCCTGAGCGGCGGTAAAAATAGTCCCGATTTAAAAATATTCATAGGTTGCGGTGGAAAACAAGTTTTTCAACTAGAATCAAGCGGATGGAGAAGCGTTTTAGATTTGCAAATAACACTACCAGCTAAAACATTGGTTTACGGAGAAATTGTTAAAGAATATCGTGGCCAAGGCAACAATCAGATGTACAGCAAATCTTTACATATTGTGGACGCACTTATGTTGGGAGGAGTTGATATCTCTGGATACCCCTTGAATGAAAGGATAAACCAATGCAACATATTTTGCACTGCCCTAAGTAAGCCCTTTGATGCTCACGTACTTCAAGTCAGATGTAAGAAGATGTTCGGGATGGAACACTTTGCTTCGGTAGTCAACAATCTCGAATACAGAGCTATGAAGCGAGTGAAGAGAGCCGTTACAATTGATGCGCCGGATCCATTGCGCAGAAACGAAATGTTTTACGTGGTTGGATCTGTGCTTTTCTTGAAAGAGATAAAAGAGCCCTGGTCATCGCAGCTATCGAAAAAAAGTGGTCTAAAATACTATTTTAGTATAGGTAAAGATAGGAAACCTCGTAGTGAATATTACATTCTGGAAGAAGCACGATTAGACATGATAGAGACATTCAGCAGTCGCCTCATGTGGTCCTGGGACGGAGCCGCTAGTGCGATACTCGATGCTCAACAACATTCCGGCCTCACGAGAGCCCAACTGGAAGAGTACATTGGTCATAATATCGATAGATAA
Protein
MNLSDTSDEGSSSEESRKPAKCSRSNTYGASSTTSHGSAIQSEDEPDTANEYHSNLSTKRRKTEIENSTDRNVRLSSRSSISENCSSPGEHFEDYRPVFNDNDEDQEENALRPSFQNSDTKFNKSVNDMNDPEYAYTGEYSEKSKRMMSNMGYKPGKGLGKFEHGRTEPVEASAQKGRRGLGLKPSIVGSVPEDFKWTPDESKPEAKEKVIWMSPAPDEPLSGDQLEEWLKKGSKKLTIDDETDFVEPHILKGVLKAKTIFDDLDDEDLRHARFRSNPYETIGSVIFLNRAAVKMANIDAVCDFVFTNPKKQNGEEVIDKKEILYFADVCAGPGGFSEYILYRRGWRAKGFGFTLKGPNDFKLSDFYAGPPETFHPYYGTKNDGNIFDPSNLMSLKEHVLQQTEDKGVHFLMADGGFSVEGQENIQEILSKQLYLCQCLAALMLVRTGGHFVVKLFDVFTRFSIGLIYIMYRCFDKVCIHKPVTSRPANSERYLICLWKRSWTESAEHHLYTVNSILWNSTNSDIDILELVPIDIIQEDTEFFEYVYKSNCILGERQIQNLLKIAAFSKHKILKEIYQAEMKEQCLQLWNLPDQVRTQPKRLNPDETLKNILSTLKTPNPHSLFNFKTNILNKKPEYIEVEGDLTNFKNVYDWHFMFLSGGKNSPDLKIFIGCGGKQVFQLESSGWRSVLDLQITLPAKTLVYGEIVKEYRGQGNNQMYSKSLHIVDALMLGGVDISGYPLNERINQCNIFCTALSKPFDAHVLQVRCKKMFGMEHFASVVNNLEYRAMKRVKRAVTIDAPDPLRRNEMFYVVGSVLFLKEIKEPWSSQLSKKSGLKYYFSIGKDRKPRSEYYILEEARLDMIETFSSRLMWSWDGAASAILDAQQHSGLTRAQLEEYIGHNIDR
Summary
Similarity
Belongs to the peptidase C2 family.
Uniprot
H9J732
A0A2H1V3E2
A0A2A4IVJ5
A0A3S2P2R4
A0A212FPQ5
A0A194QT70
+ More
A0A194PS55 A0A0L7LNS6 A0A067R9R9 A0A2J7RSM3 A0A1B3PD97 A0A336LYZ0 A0A336LYZ5 A0A336KHW6 A0A1B6C5H7 A0A232EU72 A0A182QUG1 K7IR40 T1JGN9 A0A084W1B6 A0A182XXX7 A0A2A3EUJ5 A0A1B3PDJ4 A0A088AS60 A0A3S3S4E8 A0A2R5L812 A0A158NFX7 A0A195B4D5 A0A0J7L1E6 A0A1S3K135 A0A293LBA4 A0A182M664 A0A182N1J3 A0A0N1IU74 A0A1I8MRU7 A0A182KFD8 F4W4A0 A0A182GSJ5 A0A1Z5L6E3 A0A1E1XFU5 A0A182PVZ1 A0A131XYR4 A0A182W5D9 A0A026VVC6 A0A182J3K5 A0A1B0C969 A0A131XGJ9 A0A182UQV6 A0A1Q3F634 E9H0W4 Q7PNK7 Q174B4 L7MEE7 E9IE21 A0A1B6IQL6 A0A182RMT6 A0A182TH86 A0A195E4V8 A0A195ETF6 A0A151INI9 A0A182HMM8 A0A0P5GIQ4 A0A0P6GDU0 A0A0P6I568 A0A164LGQ8 A0A0P5GFE1 E2C871 A0A131YHJ5 A0A0L7R2K9 A0A2M4AIM0 A0A151X1Q2 A0A2M4BDL1 W8C3C7 A0A0N8BWA4 A0A224YJ59 A0A1I8NLW0 A0A0P6ACV0 A0A2Y9D3T1 A0A2M4BDQ6 A0A224YJ67 A0A0P6FA20 A0A0P5N2K7 A0A0P5N204 A0A154P6Z3 A0A0N8BIF7 A0A2M4CSM1 A0A0P5DPK0 A0A2M4BEM0 A0A1E1XRJ7 A0A0P5IBQ7 A0A0P6EDG9 A0A0C9RCW8 A0A0C9QJP7 A0A1L8DGM5 A0A0K2UNK7 A0A1B0GQI7 A0A0N8DWJ1 A0A224X838 A0A023GD95 A0A0P5L8Y4
A0A194PS55 A0A0L7LNS6 A0A067R9R9 A0A2J7RSM3 A0A1B3PD97 A0A336LYZ0 A0A336LYZ5 A0A336KHW6 A0A1B6C5H7 A0A232EU72 A0A182QUG1 K7IR40 T1JGN9 A0A084W1B6 A0A182XXX7 A0A2A3EUJ5 A0A1B3PDJ4 A0A088AS60 A0A3S3S4E8 A0A2R5L812 A0A158NFX7 A0A195B4D5 A0A0J7L1E6 A0A1S3K135 A0A293LBA4 A0A182M664 A0A182N1J3 A0A0N1IU74 A0A1I8MRU7 A0A182KFD8 F4W4A0 A0A182GSJ5 A0A1Z5L6E3 A0A1E1XFU5 A0A182PVZ1 A0A131XYR4 A0A182W5D9 A0A026VVC6 A0A182J3K5 A0A1B0C969 A0A131XGJ9 A0A182UQV6 A0A1Q3F634 E9H0W4 Q7PNK7 Q174B4 L7MEE7 E9IE21 A0A1B6IQL6 A0A182RMT6 A0A182TH86 A0A195E4V8 A0A195ETF6 A0A151INI9 A0A182HMM8 A0A0P5GIQ4 A0A0P6GDU0 A0A0P6I568 A0A164LGQ8 A0A0P5GFE1 E2C871 A0A131YHJ5 A0A0L7R2K9 A0A2M4AIM0 A0A151X1Q2 A0A2M4BDL1 W8C3C7 A0A0N8BWA4 A0A224YJ59 A0A1I8NLW0 A0A0P6ACV0 A0A2Y9D3T1 A0A2M4BDQ6 A0A224YJ67 A0A0P6FA20 A0A0P5N2K7 A0A0P5N204 A0A154P6Z3 A0A0N8BIF7 A0A2M4CSM1 A0A0P5DPK0 A0A2M4BEM0 A0A1E1XRJ7 A0A0P5IBQ7 A0A0P6EDG9 A0A0C9RCW8 A0A0C9QJP7 A0A1L8DGM5 A0A0K2UNK7 A0A1B0GQI7 A0A0N8DWJ1 A0A224X838 A0A023GD95 A0A0P5L8Y4
Pubmed
EMBL
BABH01013932
ODYU01000493
SOQ35363.1
NWSH01006328
PCG63506.1
RSAL01000041
+ More
RVE50848.1 AGBW02000920 OWR55738.1 KQ461154 KPJ08514.1 KQ459595 KPI95584.1 JTDY01000457 KOB77085.1 KK852832 KDR15308.1 NEVH01000252 PNF43830.1 KU659880 AOG17679.1 UFQS01000320 UFQT01000320 SSX02836.1 SSX23204.1 SSX02837.1 SSX23205.1 SSX02835.1 SSX23203.1 GEDC01028552 JAS08746.1 NNAY01002190 OXU21846.1 AXCN02001449 JH432211 ATLV01019258 KE525266 KFB44010.1 KZ288186 PBC34809.1 KU660003 AOG17801.1 NCKU01003052 NCKU01002705 RWS08274.1 RWS08968.1 GGLE01001469 MBY05595.1 ADTU01001538 KQ976625 KYM79059.1 LBMM01001287 KMQ96577.1 GFWV01000338 MAA25068.1 AXCM01000181 KQ435693 KOX81002.1 GL887491 EGI71096.1 JXUM01084758 KQ563457 KXJ73806.1 GFJQ02004105 JAW02865.1 GFAC01001232 JAT97956.1 GEFM01004384 JAP71412.1 KK107847 QOIP01000005 EZA47486.1 RLU23040.1 AJWK01001964 GEFH01002894 JAP65687.1 GFDL01012011 JAV23034.1 GL732582 EFX74631.1 AAAB01008963 EAA12036.6 CH477414 EAT41411.1 GACK01002659 JAA62375.1 GL762556 EFZ21153.1 GECU01018497 JAS89209.1 KQ979609 KYN20198.1 KQ981979 KYN31441.1 KQ976932 KYN07037.1 APCN01004841 APCN01004842 GDIQ01239982 JAK11743.1 GDIQ01034606 JAN60131.1 GDIQ01016744 JAN77993.1 LRGB01003134 KZS04093.1 GDIQ01245426 JAK06299.1 GL453568 EFN75874.1 GEDV01009803 JAP78754.1 KQ414666 KOC65112.1 GGFK01007309 MBW40630.1 KQ982585 KYQ54327.1 GGFJ01001993 MBW51134.1 GAMC01002782 JAC03774.1 GDIQ01138627 JAL13099.1 GFPF01006542 MAA17688.1 GDIP01036149 JAM67566.1 GGFJ01001992 MBW51133.1 GFPF01006541 MAA17687.1 GDIQ01050807 JAN43930.1 GDIQ01148224 JAL03502.1 GDIQ01148225 JAL03501.1 KQ434829 KZC07705.1 GDIQ01174603 JAK77122.1 GGFL01004166 MBW68344.1 GDIP01156731 JAJ66671.1 GGFJ01002302 MBW51443.1 GFAA01001494 JAU01941.1 GDIQ01215693 JAK36032.1 GDIQ01064347 JAN30390.1 GBYB01004741 JAG74508.1 GBYB01000782 GBYB01000784 JAG70549.1 JAG70551.1 GFDF01008476 JAV05608.1 HACA01022472 CDW39833.1 AJVK01007261 GDIQ01084940 JAN09797.1 GFTR01007910 JAW08516.1 GBBM01003591 JAC31827.1 GDIQ01178346 JAK73379.1
RVE50848.1 AGBW02000920 OWR55738.1 KQ461154 KPJ08514.1 KQ459595 KPI95584.1 JTDY01000457 KOB77085.1 KK852832 KDR15308.1 NEVH01000252 PNF43830.1 KU659880 AOG17679.1 UFQS01000320 UFQT01000320 SSX02836.1 SSX23204.1 SSX02837.1 SSX23205.1 SSX02835.1 SSX23203.1 GEDC01028552 JAS08746.1 NNAY01002190 OXU21846.1 AXCN02001449 JH432211 ATLV01019258 KE525266 KFB44010.1 KZ288186 PBC34809.1 KU660003 AOG17801.1 NCKU01003052 NCKU01002705 RWS08274.1 RWS08968.1 GGLE01001469 MBY05595.1 ADTU01001538 KQ976625 KYM79059.1 LBMM01001287 KMQ96577.1 GFWV01000338 MAA25068.1 AXCM01000181 KQ435693 KOX81002.1 GL887491 EGI71096.1 JXUM01084758 KQ563457 KXJ73806.1 GFJQ02004105 JAW02865.1 GFAC01001232 JAT97956.1 GEFM01004384 JAP71412.1 KK107847 QOIP01000005 EZA47486.1 RLU23040.1 AJWK01001964 GEFH01002894 JAP65687.1 GFDL01012011 JAV23034.1 GL732582 EFX74631.1 AAAB01008963 EAA12036.6 CH477414 EAT41411.1 GACK01002659 JAA62375.1 GL762556 EFZ21153.1 GECU01018497 JAS89209.1 KQ979609 KYN20198.1 KQ981979 KYN31441.1 KQ976932 KYN07037.1 APCN01004841 APCN01004842 GDIQ01239982 JAK11743.1 GDIQ01034606 JAN60131.1 GDIQ01016744 JAN77993.1 LRGB01003134 KZS04093.1 GDIQ01245426 JAK06299.1 GL453568 EFN75874.1 GEDV01009803 JAP78754.1 KQ414666 KOC65112.1 GGFK01007309 MBW40630.1 KQ982585 KYQ54327.1 GGFJ01001993 MBW51134.1 GAMC01002782 JAC03774.1 GDIQ01138627 JAL13099.1 GFPF01006542 MAA17688.1 GDIP01036149 JAM67566.1 GGFJ01001992 MBW51133.1 GFPF01006541 MAA17687.1 GDIQ01050807 JAN43930.1 GDIQ01148224 JAL03502.1 GDIQ01148225 JAL03501.1 KQ434829 KZC07705.1 GDIQ01174603 JAK77122.1 GGFL01004166 MBW68344.1 GDIP01156731 JAJ66671.1 GGFJ01002302 MBW51443.1 GFAA01001494 JAU01941.1 GDIQ01215693 JAK36032.1 GDIQ01064347 JAN30390.1 GBYB01004741 JAG74508.1 GBYB01000782 GBYB01000784 JAG70549.1 JAG70551.1 GFDF01008476 JAV05608.1 HACA01022472 CDW39833.1 AJVK01007261 GDIQ01084940 JAN09797.1 GFTR01007910 JAW08516.1 GBBM01003591 JAC31827.1 GDIQ01178346 JAK73379.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
+ More
UP000037510 UP000027135 UP000235965 UP000215335 UP000075886 UP000002358 UP000030765 UP000076408 UP000242457 UP000005203 UP000285301 UP000005205 UP000078540 UP000036403 UP000085678 UP000075883 UP000075884 UP000053105 UP000095301 UP000075881 UP000007755 UP000069940 UP000249989 UP000075885 UP000075920 UP000053097 UP000279307 UP000075880 UP000092461 UP000075903 UP000000305 UP000007062 UP000008820 UP000075900 UP000075902 UP000078492 UP000078541 UP000078542 UP000075840 UP000076858 UP000008237 UP000053825 UP000075809 UP000095300 UP000076407 UP000076502 UP000092462
UP000037510 UP000027135 UP000235965 UP000215335 UP000075886 UP000002358 UP000030765 UP000076408 UP000242457 UP000005203 UP000285301 UP000005205 UP000078540 UP000036403 UP000085678 UP000075883 UP000075884 UP000053105 UP000095301 UP000075881 UP000007755 UP000069940 UP000249989 UP000075885 UP000075920 UP000053097 UP000279307 UP000075880 UP000092461 UP000075903 UP000000305 UP000007062 UP000008820 UP000075900 UP000075902 UP000078492 UP000078541 UP000078542 UP000075840 UP000076858 UP000008237 UP000053825 UP000075809 UP000095300 UP000076407 UP000076502 UP000092462
PRIDE
Pfam
Interpro
IPR029063
SAM-dependent_MTases
+ More
IPR025816 RrmJ-type_MeTrfase
IPR002877 rRNA_MeTrfase_FtsJ_dom
IPR030376 Cap_mRNA_MeTrfase_1
IPR000467 G_patch_dom
IPR003593 AAA+_ATPase
IPR003959 ATPase_AAA_core
IPR027417 P-loop_NTPase
IPR011417 ANTH_dom
IPR013809 ENTH
IPR008942 ENTH_VHS
IPR014712 Clathrin_AP_dom2
IPR001202 WW_dom
IPR036020 WW_dom_sf
IPR036372 BEACH_dom_sf
IPR001452 SH3_domain
IPR001605 PH_dom-spectrin-type
IPR001715 CH-domain
IPR036872 CH_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR018159 Spectrin/alpha-actinin
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR002017 Spectrin_repeat
IPR011993 PH-like_dom_sf
IPR041681 PH_9
IPR001589 Actinin_actin-bd_CS
IPR036322 WD40_repeat_dom_sf
IPR000409 BEACH_dom
IPR011009 Kinase-like_dom_sf
IPR036028 SH3-like_dom_sf
IPR001849 PH_domain
IPR000169 Pept_cys_AS
IPR001300 Peptidase_C2_calpain_cat
IPR001876 Znf_RanBP2
IPR038765 Papain-like_cys_pep_sf
IPR022684 Calpain_cysteine_protease
IPR014014 RNA_helicase_DEAD_Q_motif
IPR011545 DEAD/DEAH_box_helicase_dom
IPR000629 RNA-helicase_DEAD-box_CS
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR025816 RrmJ-type_MeTrfase
IPR002877 rRNA_MeTrfase_FtsJ_dom
IPR030376 Cap_mRNA_MeTrfase_1
IPR000467 G_patch_dom
IPR003593 AAA+_ATPase
IPR003959 ATPase_AAA_core
IPR027417 P-loop_NTPase
IPR011417 ANTH_dom
IPR013809 ENTH
IPR008942 ENTH_VHS
IPR014712 Clathrin_AP_dom2
IPR001202 WW_dom
IPR036020 WW_dom_sf
IPR036372 BEACH_dom_sf
IPR001452 SH3_domain
IPR001605 PH_dom-spectrin-type
IPR001715 CH-domain
IPR036872 CH_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR018159 Spectrin/alpha-actinin
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR002017 Spectrin_repeat
IPR011993 PH-like_dom_sf
IPR041681 PH_9
IPR001589 Actinin_actin-bd_CS
IPR036322 WD40_repeat_dom_sf
IPR000409 BEACH_dom
IPR011009 Kinase-like_dom_sf
IPR036028 SH3-like_dom_sf
IPR001849 PH_domain
IPR000169 Pept_cys_AS
IPR001300 Peptidase_C2_calpain_cat
IPR001876 Znf_RanBP2
IPR038765 Papain-like_cys_pep_sf
IPR022684 Calpain_cysteine_protease
IPR014014 RNA_helicase_DEAD_Q_motif
IPR011545 DEAD/DEAH_box_helicase_dom
IPR000629 RNA-helicase_DEAD-box_CS
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
SUPFAM
ProteinModelPortal
H9J732
A0A2H1V3E2
A0A2A4IVJ5
A0A3S2P2R4
A0A212FPQ5
A0A194QT70
+ More
A0A194PS55 A0A0L7LNS6 A0A067R9R9 A0A2J7RSM3 A0A1B3PD97 A0A336LYZ0 A0A336LYZ5 A0A336KHW6 A0A1B6C5H7 A0A232EU72 A0A182QUG1 K7IR40 T1JGN9 A0A084W1B6 A0A182XXX7 A0A2A3EUJ5 A0A1B3PDJ4 A0A088AS60 A0A3S3S4E8 A0A2R5L812 A0A158NFX7 A0A195B4D5 A0A0J7L1E6 A0A1S3K135 A0A293LBA4 A0A182M664 A0A182N1J3 A0A0N1IU74 A0A1I8MRU7 A0A182KFD8 F4W4A0 A0A182GSJ5 A0A1Z5L6E3 A0A1E1XFU5 A0A182PVZ1 A0A131XYR4 A0A182W5D9 A0A026VVC6 A0A182J3K5 A0A1B0C969 A0A131XGJ9 A0A182UQV6 A0A1Q3F634 E9H0W4 Q7PNK7 Q174B4 L7MEE7 E9IE21 A0A1B6IQL6 A0A182RMT6 A0A182TH86 A0A195E4V8 A0A195ETF6 A0A151INI9 A0A182HMM8 A0A0P5GIQ4 A0A0P6GDU0 A0A0P6I568 A0A164LGQ8 A0A0P5GFE1 E2C871 A0A131YHJ5 A0A0L7R2K9 A0A2M4AIM0 A0A151X1Q2 A0A2M4BDL1 W8C3C7 A0A0N8BWA4 A0A224YJ59 A0A1I8NLW0 A0A0P6ACV0 A0A2Y9D3T1 A0A2M4BDQ6 A0A224YJ67 A0A0P6FA20 A0A0P5N2K7 A0A0P5N204 A0A154P6Z3 A0A0N8BIF7 A0A2M4CSM1 A0A0P5DPK0 A0A2M4BEM0 A0A1E1XRJ7 A0A0P5IBQ7 A0A0P6EDG9 A0A0C9RCW8 A0A0C9QJP7 A0A1L8DGM5 A0A0K2UNK7 A0A1B0GQI7 A0A0N8DWJ1 A0A224X838 A0A023GD95 A0A0P5L8Y4
A0A194PS55 A0A0L7LNS6 A0A067R9R9 A0A2J7RSM3 A0A1B3PD97 A0A336LYZ0 A0A336LYZ5 A0A336KHW6 A0A1B6C5H7 A0A232EU72 A0A182QUG1 K7IR40 T1JGN9 A0A084W1B6 A0A182XXX7 A0A2A3EUJ5 A0A1B3PDJ4 A0A088AS60 A0A3S3S4E8 A0A2R5L812 A0A158NFX7 A0A195B4D5 A0A0J7L1E6 A0A1S3K135 A0A293LBA4 A0A182M664 A0A182N1J3 A0A0N1IU74 A0A1I8MRU7 A0A182KFD8 F4W4A0 A0A182GSJ5 A0A1Z5L6E3 A0A1E1XFU5 A0A182PVZ1 A0A131XYR4 A0A182W5D9 A0A026VVC6 A0A182J3K5 A0A1B0C969 A0A131XGJ9 A0A182UQV6 A0A1Q3F634 E9H0W4 Q7PNK7 Q174B4 L7MEE7 E9IE21 A0A1B6IQL6 A0A182RMT6 A0A182TH86 A0A195E4V8 A0A195ETF6 A0A151INI9 A0A182HMM8 A0A0P5GIQ4 A0A0P6GDU0 A0A0P6I568 A0A164LGQ8 A0A0P5GFE1 E2C871 A0A131YHJ5 A0A0L7R2K9 A0A2M4AIM0 A0A151X1Q2 A0A2M4BDL1 W8C3C7 A0A0N8BWA4 A0A224YJ59 A0A1I8NLW0 A0A0P6ACV0 A0A2Y9D3T1 A0A2M4BDQ6 A0A224YJ67 A0A0P6FA20 A0A0P5N2K7 A0A0P5N204 A0A154P6Z3 A0A0N8BIF7 A0A2M4CSM1 A0A0P5DPK0 A0A2M4BEM0 A0A1E1XRJ7 A0A0P5IBQ7 A0A0P6EDG9 A0A0C9RCW8 A0A0C9QJP7 A0A1L8DGM5 A0A0K2UNK7 A0A1B0GQI7 A0A0N8DWJ1 A0A224X838 A0A023GD95 A0A0P5L8Y4
PDB
4N4A
E-value=3.49111e-120,
Score=1108
Ontologies
GO
PANTHER
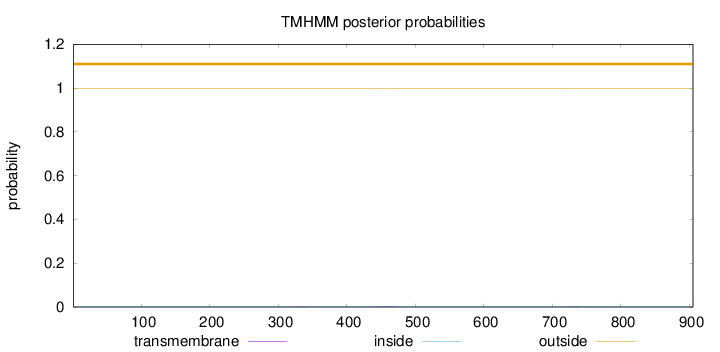
Topology
Length:
905
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0616800000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00066
outside
1 - 905
Population Genetic Test Statistics
Pi
211.929652
Theta
152.104059
Tajima's D
1.208216
CLR
0.162083
CSRT
0.72096395180241
Interpretation
Uncertain
