Gene
KWMTBOMO04587 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005485
Annotation
PREDICTED:_ADP-ribose_pyrophosphatase?_mitochondrial_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 0.995
Sequence
CDS
ATGAATTCAATATTAACTGTGCATTCCAAATGTCGTGTAGGATTTTATCCTAGGAGTAATTTAAAAAGATTTCCTGTTCCAGATGACAAAGTCGACTGGTCTACCGAGTATAAACAATATAATCCTCCTAATTACACATTTCCAGGCTTACATGGAAAACCATATGCAGATCCAGAAATCGGTGATCCTAACTTTGAACCTTTATGGAATAGCATTGATGGCAATATAAGTAGAGTGAGTTACAATGGACCATATCAAATTGTCAATGGTTTCCCTTTAAACCCTCTGGGACGTACTGGTATCTGTGGTCGAGGTGTTTTAGGACGATGGGGTCCTAATCATGCTGCAGACCCAATTATAAGTCGTTGGAAAAGACTTGACAATGGAAATATGGCTGTGGGTGTAAATAATAAACCTATATTACAATTTATAGCTATAAAACGGGGTGACACTGGAGAGTGGGCTATACCAGGTGGTATGGTGGATCCTGGTGAAAAAGTTAGTGAGACTGCTATAAGAGAATTTAAAGAAGAAGCTCTTAATTCATTAAGCCTGGCAGCCTGCTTTTCAAACCCCATTTGA
Protein
MNSILTVHSKCRVGFYPRSNLKRFPVPDDKVDWSTEYKQYNPPNYTFPGLHGKPYADPEIGDPNFEPLWNSIDGNISRVSYNGPYQIVNGFPLNPLGRTGICGRGVLGRWGPNHAADPIISRWKRLDNGNMAVGVNNKPILQFIAIKRGDTGEWAIPGGMVDPGEKVSETAIREFKEEALNSLSLAACFSNPI
Summary
Uniprot
H9J7J3
A0A2A4IXN3
A0A2A4IW37
A0A194QSK7
A0A212EL76
A0A2H1W582
+ More
A0A154PDJ8 A0A2P8YAJ7 A0A0A1WUZ5 A0A1B6JXX1 A0A1B6I8Q6 E2AKJ8 A0A1B6EWL6 R7UTP2 A0A0L7R965 A0A2J7R707 A0A131XIH7 A0A195F404 A0A067R445 A0A1W4W4K9 A0A195DVB1 A0A151I828 R7T9B1 A0A224YQV3 A0A131YC05 A0A026WPZ0 A0A3L8DNC8 A0A224XPN0 A0A034WP32 F4WIM3 A0A0V0GA23 A0A0J7KXS4 A0A310SCL2 A0A2I4B3K2 L7M4R6 A0A131YHX9 A0A069DRH2 A0A1E1XFN3 A0A1Z5LGS7 A0A151XE20 A0A023F7K5 A0A293LKA9 A0A0M9A1R1 A0A195BHK5 A0A023GHX9 A0A2A3EAB1 A0A0V1DJS2 A0A232F156 G3MM24 A0A2I4B3K0 A0A2R7W7L4 A0A3B5A0H1 K7J3L0 A0A1E1XRE0 A0A1Y3EK96 A0A0V0UD74 A0A0V1PNW6 E2BWY5 A0A1B6M1F3 A0A1A8BU66 A0A1A8E2Z9 A0A0V0VZX4 A0A1A8JF00 D6WVF4 A0A1A8K9G5 A0A0V0X5K7 A0A3Q2PWB2 A0A1A8EMJ2 A0A2S2Q3E1 A0A0V1ADX4 A0A315WBV5 A0A3Q1GM62 A0A146UWZ8 A0A3Q3KGW6 A0A3Q1FFI8 A0A3Q0REZ9 G3PID6 A0A3N0Y274 A0A3Q1BLX8 A0A3P8TBV7 A0A1A8A9V3 A0A1A8LDQ7 A0A0F8ALD8 A0A3Q0RF09 A0A1I8NHG4 A0A0V1MUU2 A0A146ZK05 A0A3Q1ISK5 A0A1A8REN5 A0A3Q1HP02 A0A1A8NF32 A0A0V1I0J8 A0A1B6DPX1 A0A3B4UVT5 A0A182FHM0 A7MCK5 A0A1Y1N1E9 F1QL34 Q6PFU2 A0A3B4YP16
A0A154PDJ8 A0A2P8YAJ7 A0A0A1WUZ5 A0A1B6JXX1 A0A1B6I8Q6 E2AKJ8 A0A1B6EWL6 R7UTP2 A0A0L7R965 A0A2J7R707 A0A131XIH7 A0A195F404 A0A067R445 A0A1W4W4K9 A0A195DVB1 A0A151I828 R7T9B1 A0A224YQV3 A0A131YC05 A0A026WPZ0 A0A3L8DNC8 A0A224XPN0 A0A034WP32 F4WIM3 A0A0V0GA23 A0A0J7KXS4 A0A310SCL2 A0A2I4B3K2 L7M4R6 A0A131YHX9 A0A069DRH2 A0A1E1XFN3 A0A1Z5LGS7 A0A151XE20 A0A023F7K5 A0A293LKA9 A0A0M9A1R1 A0A195BHK5 A0A023GHX9 A0A2A3EAB1 A0A0V1DJS2 A0A232F156 G3MM24 A0A2I4B3K0 A0A2R7W7L4 A0A3B5A0H1 K7J3L0 A0A1E1XRE0 A0A1Y3EK96 A0A0V0UD74 A0A0V1PNW6 E2BWY5 A0A1B6M1F3 A0A1A8BU66 A0A1A8E2Z9 A0A0V0VZX4 A0A1A8JF00 D6WVF4 A0A1A8K9G5 A0A0V0X5K7 A0A3Q2PWB2 A0A1A8EMJ2 A0A2S2Q3E1 A0A0V1ADX4 A0A315WBV5 A0A3Q1GM62 A0A146UWZ8 A0A3Q3KGW6 A0A3Q1FFI8 A0A3Q0REZ9 G3PID6 A0A3N0Y274 A0A3Q1BLX8 A0A3P8TBV7 A0A1A8A9V3 A0A1A8LDQ7 A0A0F8ALD8 A0A3Q0RF09 A0A1I8NHG4 A0A0V1MUU2 A0A146ZK05 A0A3Q1ISK5 A0A1A8REN5 A0A3Q1HP02 A0A1A8NF32 A0A0V1I0J8 A0A1B6DPX1 A0A3B4UVT5 A0A182FHM0 A7MCK5 A0A1Y1N1E9 F1QL34 Q6PFU2 A0A3B4YP16
Pubmed
EMBL
BABH01013925
NWSH01005707
PCG64024.1
PCG64025.1
KQ461154
KPJ08508.1
+ More
AGBW02014132 OWR42221.1 ODYU01006382 SOQ48183.1 KQ434879 KZC09975.1 PYGN01000754 PSN41282.1 GBXI01011418 JAD02874.1 GECU01003677 JAT04030.1 GECU01024414 JAS83292.1 GL440281 EFN66049.1 GECZ01027431 JAS42338.1 AMQN01022379 KB300039 ELU07292.1 KQ414628 KOC67331.1 NEVH01006738 PNF36628.1 GEFH01002633 JAP65948.1 KQ981820 KYN35310.1 KK852994 KDR12722.1 KQ980304 KYN16786.1 KQ978390 KYM94301.1 AMQN01003598 KB312243 ELT87584.1 GFPF01005825 MAA16971.1 GEFM01000180 JAP75616.1 KK107135 EZA58090.1 QOIP01000006 RLU21318.1 GFTR01004638 JAW11788.1 GAKP01002865 JAC56087.1 GL888176 EGI65959.1 GECL01001148 JAP04976.1 LBMM01002043 KMQ95357.1 KQ761323 OAD57903.1 GACK01005703 JAA59331.1 GEDV01010000 JAP78557.1 GBGD01002384 JAC86505.1 GFAC01001136 JAT98052.1 GFJQ02000357 JAW06613.1 KQ982254 KYQ58626.1 GBBI01001495 JAC17217.1 GFWV01004194 MAA28924.1 KQ435794 KOX74019.1 KQ976465 KYM84303.1 GBBM01001907 JAC33511.1 KZ288309 PBC28667.1 JYDI01000001 KRY61560.1 NNAY01001394 OXU24128.1 JO842925 AEO34542.1 KK854407 PTY15569.1 GFAA01001943 JAU01492.1 LVZM01008977 OUC45592.1 JYDJ01000019 KRX49202.1 JYDM01000001 KRZ97939.1 GL451188 EFN79849.1 GEBQ01010208 GEBQ01001330 JAT29769.1 JAT38647.1 HADZ01007271 SBP71212.1 HAEA01011391 SBQ39871.1 JYDN01000001 KRX69023.1 HAED01020908 SBR07543.1 KQ971357 EFA08307.1 HAEE01008264 SBR28314.1 JYDK01000015 KRX83188.1 HAEB01000527 HAEC01011262 SBQ46975.1 GGMS01003042 MBY72245.1 JYDQ01000009 KRY22419.1 NHOQ01000074 PWA33235.1 GCES01076805 JAR09518.1 RJVU01053643 ROL27973.1 HADY01012495 HAEJ01010995 SBP50980.1 HAEF01004617 SBR41999.1 KQ041555 KKF25583.1 JYDO01000037 KRZ75554.1 GCES01020126 JAR66197.1 HAEI01007997 SBS03853.1 HAEH01001721 SBR67469.1 JYDP01000012 KRZ16381.1 GEDC01009563 JAS27735.1 BC152216 AAI52217.1 GEZM01017740 JAV90485.1 FP016144 BC057417 AAH57417.1
AGBW02014132 OWR42221.1 ODYU01006382 SOQ48183.1 KQ434879 KZC09975.1 PYGN01000754 PSN41282.1 GBXI01011418 JAD02874.1 GECU01003677 JAT04030.1 GECU01024414 JAS83292.1 GL440281 EFN66049.1 GECZ01027431 JAS42338.1 AMQN01022379 KB300039 ELU07292.1 KQ414628 KOC67331.1 NEVH01006738 PNF36628.1 GEFH01002633 JAP65948.1 KQ981820 KYN35310.1 KK852994 KDR12722.1 KQ980304 KYN16786.1 KQ978390 KYM94301.1 AMQN01003598 KB312243 ELT87584.1 GFPF01005825 MAA16971.1 GEFM01000180 JAP75616.1 KK107135 EZA58090.1 QOIP01000006 RLU21318.1 GFTR01004638 JAW11788.1 GAKP01002865 JAC56087.1 GL888176 EGI65959.1 GECL01001148 JAP04976.1 LBMM01002043 KMQ95357.1 KQ761323 OAD57903.1 GACK01005703 JAA59331.1 GEDV01010000 JAP78557.1 GBGD01002384 JAC86505.1 GFAC01001136 JAT98052.1 GFJQ02000357 JAW06613.1 KQ982254 KYQ58626.1 GBBI01001495 JAC17217.1 GFWV01004194 MAA28924.1 KQ435794 KOX74019.1 KQ976465 KYM84303.1 GBBM01001907 JAC33511.1 KZ288309 PBC28667.1 JYDI01000001 KRY61560.1 NNAY01001394 OXU24128.1 JO842925 AEO34542.1 KK854407 PTY15569.1 GFAA01001943 JAU01492.1 LVZM01008977 OUC45592.1 JYDJ01000019 KRX49202.1 JYDM01000001 KRZ97939.1 GL451188 EFN79849.1 GEBQ01010208 GEBQ01001330 JAT29769.1 JAT38647.1 HADZ01007271 SBP71212.1 HAEA01011391 SBQ39871.1 JYDN01000001 KRX69023.1 HAED01020908 SBR07543.1 KQ971357 EFA08307.1 HAEE01008264 SBR28314.1 JYDK01000015 KRX83188.1 HAEB01000527 HAEC01011262 SBQ46975.1 GGMS01003042 MBY72245.1 JYDQ01000009 KRY22419.1 NHOQ01000074 PWA33235.1 GCES01076805 JAR09518.1 RJVU01053643 ROL27973.1 HADY01012495 HAEJ01010995 SBP50980.1 HAEF01004617 SBR41999.1 KQ041555 KKF25583.1 JYDO01000037 KRZ75554.1 GCES01020126 JAR66197.1 HAEI01007997 SBS03853.1 HAEH01001721 SBR67469.1 JYDP01000012 KRZ16381.1 GEDC01009563 JAS27735.1 BC152216 AAI52217.1 GEZM01017740 JAV90485.1 FP016144 BC057417 AAH57417.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000076502
UP000245037
+ More
UP000000311 UP000014760 UP000053825 UP000235965 UP000078541 UP000027135 UP000192223 UP000078492 UP000078542 UP000053097 UP000279307 UP000007755 UP000036403 UP000192220 UP000075809 UP000053105 UP000078540 UP000242457 UP000054653 UP000215335 UP000261400 UP000002358 UP000243006 UP000055048 UP000054924 UP000008237 UP000054681 UP000007266 UP000054673 UP000265000 UP000054783 UP000257200 UP000261600 UP000261340 UP000007635 UP000257160 UP000265080 UP000095301 UP000054843 UP000265040 UP000055024 UP000261420 UP000069272 UP000000437 UP000261360
UP000000311 UP000014760 UP000053825 UP000235965 UP000078541 UP000027135 UP000192223 UP000078492 UP000078542 UP000053097 UP000279307 UP000007755 UP000036403 UP000192220 UP000075809 UP000053105 UP000078540 UP000242457 UP000054653 UP000215335 UP000261400 UP000002358 UP000243006 UP000055048 UP000054924 UP000008237 UP000054681 UP000007266 UP000054673 UP000265000 UP000054783 UP000257200 UP000261600 UP000261340 UP000007635 UP000257160 UP000265080 UP000095301 UP000054843 UP000265040 UP000055024 UP000261420 UP000069272 UP000000437 UP000261360
Interpro
Gene 3D
ProteinModelPortal
H9J7J3
A0A2A4IXN3
A0A2A4IW37
A0A194QSK7
A0A212EL76
A0A2H1W582
+ More
A0A154PDJ8 A0A2P8YAJ7 A0A0A1WUZ5 A0A1B6JXX1 A0A1B6I8Q6 E2AKJ8 A0A1B6EWL6 R7UTP2 A0A0L7R965 A0A2J7R707 A0A131XIH7 A0A195F404 A0A067R445 A0A1W4W4K9 A0A195DVB1 A0A151I828 R7T9B1 A0A224YQV3 A0A131YC05 A0A026WPZ0 A0A3L8DNC8 A0A224XPN0 A0A034WP32 F4WIM3 A0A0V0GA23 A0A0J7KXS4 A0A310SCL2 A0A2I4B3K2 L7M4R6 A0A131YHX9 A0A069DRH2 A0A1E1XFN3 A0A1Z5LGS7 A0A151XE20 A0A023F7K5 A0A293LKA9 A0A0M9A1R1 A0A195BHK5 A0A023GHX9 A0A2A3EAB1 A0A0V1DJS2 A0A232F156 G3MM24 A0A2I4B3K0 A0A2R7W7L4 A0A3B5A0H1 K7J3L0 A0A1E1XRE0 A0A1Y3EK96 A0A0V0UD74 A0A0V1PNW6 E2BWY5 A0A1B6M1F3 A0A1A8BU66 A0A1A8E2Z9 A0A0V0VZX4 A0A1A8JF00 D6WVF4 A0A1A8K9G5 A0A0V0X5K7 A0A3Q2PWB2 A0A1A8EMJ2 A0A2S2Q3E1 A0A0V1ADX4 A0A315WBV5 A0A3Q1GM62 A0A146UWZ8 A0A3Q3KGW6 A0A3Q1FFI8 A0A3Q0REZ9 G3PID6 A0A3N0Y274 A0A3Q1BLX8 A0A3P8TBV7 A0A1A8A9V3 A0A1A8LDQ7 A0A0F8ALD8 A0A3Q0RF09 A0A1I8NHG4 A0A0V1MUU2 A0A146ZK05 A0A3Q1ISK5 A0A1A8REN5 A0A3Q1HP02 A0A1A8NF32 A0A0V1I0J8 A0A1B6DPX1 A0A3B4UVT5 A0A182FHM0 A7MCK5 A0A1Y1N1E9 F1QL34 Q6PFU2 A0A3B4YP16
A0A154PDJ8 A0A2P8YAJ7 A0A0A1WUZ5 A0A1B6JXX1 A0A1B6I8Q6 E2AKJ8 A0A1B6EWL6 R7UTP2 A0A0L7R965 A0A2J7R707 A0A131XIH7 A0A195F404 A0A067R445 A0A1W4W4K9 A0A195DVB1 A0A151I828 R7T9B1 A0A224YQV3 A0A131YC05 A0A026WPZ0 A0A3L8DNC8 A0A224XPN0 A0A034WP32 F4WIM3 A0A0V0GA23 A0A0J7KXS4 A0A310SCL2 A0A2I4B3K2 L7M4R6 A0A131YHX9 A0A069DRH2 A0A1E1XFN3 A0A1Z5LGS7 A0A151XE20 A0A023F7K5 A0A293LKA9 A0A0M9A1R1 A0A195BHK5 A0A023GHX9 A0A2A3EAB1 A0A0V1DJS2 A0A232F156 G3MM24 A0A2I4B3K0 A0A2R7W7L4 A0A3B5A0H1 K7J3L0 A0A1E1XRE0 A0A1Y3EK96 A0A0V0UD74 A0A0V1PNW6 E2BWY5 A0A1B6M1F3 A0A1A8BU66 A0A1A8E2Z9 A0A0V0VZX4 A0A1A8JF00 D6WVF4 A0A1A8K9G5 A0A0V0X5K7 A0A3Q2PWB2 A0A1A8EMJ2 A0A2S2Q3E1 A0A0V1ADX4 A0A315WBV5 A0A3Q1GM62 A0A146UWZ8 A0A3Q3KGW6 A0A3Q1FFI8 A0A3Q0REZ9 G3PID6 A0A3N0Y274 A0A3Q1BLX8 A0A3P8TBV7 A0A1A8A9V3 A0A1A8LDQ7 A0A0F8ALD8 A0A3Q0RF09 A0A1I8NHG4 A0A0V1MUU2 A0A146ZK05 A0A3Q1ISK5 A0A1A8REN5 A0A3Q1HP02 A0A1A8NF32 A0A0V1I0J8 A0A1B6DPX1 A0A3B4UVT5 A0A182FHM0 A7MCK5 A0A1Y1N1E9 F1QL34 Q6PFU2 A0A3B4YP16
PDB
1QVJ
E-value=1.53068e-53,
Score=526
Ontologies
PATHWAY
GO
PANTHER
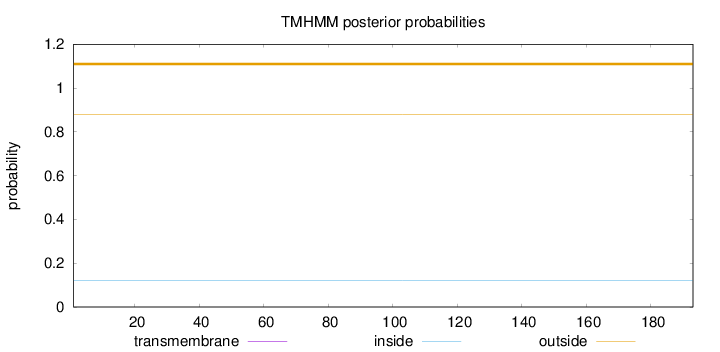
Topology
Length:
193
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0015
Exp number, first 60 AAs:
0
Total prob of N-in:
0.11954
outside
1 - 193
Population Genetic Test Statistics
Pi
124.32797
Theta
143.621615
Tajima's D
-0.366567
CLR
1.136279
CSRT
0.272436378181091
Interpretation
Uncertain
