Gene
KWMTBOMO04586
Pre Gene Modal
BGIBMGA005484
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_condensin_complex_subunit_1_[Bombyx_mori]
Full name
Condensin complex subunit 1
Location in the cell
Nuclear Reliability : 3.707
Sequence
CDS
ATGATTACGTTACTCAAAAGATCGAAAAACGTATCTTTGAGAACCAATTTGACAATTGCCTTCGCCGACCTCACTTTGAGATTCCCGAATCTTACGCAGCCGTGGACGCATCACATTTATCACATTCTAAGCGACGAGGAGCTAGAAGTCCGTCAGTGTGCAGTGAAGATGCTGTCGTTTCTGGTCCTGCACGAGATGGTCCGCGTGAAGGGACAGATAGCAGACATGGCGCTGTGCTGCGCGGACGCGGACGCTAAAGTGGCGAGTATGACGCGCCTCTTCTTCAAACAGCTTTCACAAAAGGGAAACGCCCTATACAACGTCATGCCCGATATTATATCAAGACTGAGCGATCCGGATTTGAATGTACCTGAAGAGCAATACAGAATTATTATGAAATATATAACTTCACTAATCCAAAAAGACAGACAAATGGAGGCACTAGTAGAGAAATTATGCCAAAGATTCAAATTATCAACAGAAGAAAGACAGTGGCGAGACTTAGCTTATTGTCTCTCTCTGTTTACATACAACGAAAGGTCACTAAGGAAACTCATTGAAAATCTCGACTGTTACAAAGATAAATTATATTGTCAAGGAGTGATGGAGTCTTTCAACACAATTATGAATAACGCTTCCAAAATGGCCAAGAACGACATTAAGGCTTTGGTAACCGAACTGGGAGATAAAATAGAAGAAGGCTTTGCGGTTCGCGCTGAGGAAGGTAATTGTGAAGGAACCACTGAAGGCAGTGGCGACACTGACAAGAAGGCGACGGGTCCGCGCGCCACTCCCAGACGGCGGGCTGCGCCCCGAAGGAACCGCAGGAGATCTTCATCCAGTGCAGATGAGAATGAACCTCCCAGCAACGTCGAAACCCCAGTGTCGACTAGAAAATCTAATCGGAAAGCAGCAAATAAACAAAAGATTGTAAATGATTCAGAAGATTCAGATGACGATGACTTTCAGGATAAAGAGGAAGTGTTTAAAAAACCAGTGAGAAAAGCAACGCGACGAAGAAAGTTATAA
Protein
MITLLKRSKNVSLRTNLTIAFADLTLRFPNLTQPWTHHIYHILSDEELEVRQCAVKMLSFLVLHEMVRVKGQIADMALCCADADAKVASMTRLFFKQLSQKGNALYNVMPDIISRLSDPDLNVPEEQYRIIMKYITSLIQKDRQMEALVEKLCQRFKLSTEERQWRDLAYCLSLFTYNERSLRKLIENLDCYKDKLYCQGVMESFNTIMNNASKMAKNDIKALVTELGDKIEEGFAVRAEEGNCEGTTEGSGDTDKKATGPRATPRRRAAPRRNRRRSSSSADENEPPSNVETPVSTRKSNRKAANKQKIVNDSEDSDDDDFQDKEEVFKKPVRKATRRRKL
Summary
Description
Regulatory subunit of the condensin complex, a complex required for conversion of interphase chromatin into mitotic-like condense chromosomes. The condensin complex probably introduces positive supercoils into relaxed DNA in the presence of type I topoisomerases and converts nicked DNA into positive knotted forms in the presence of type II topoisomerases.
Similarity
Belongs to the CND1 (condensin subunit 1) family.
Uniprot
H9J7J2
S4NYG7
A0A2H1W582
A0A3S2NV77
A0A212EL47
A0A194QSP9
+ More
A0A194PQ56 A0A0L7KRA2 A0A1I8PTP0 A0A1I8PTS8 A0A1I8MCN6 W8CBD0 A0A182MHJ2 A0A182VSA9 A0A182HJT6 Q7QDI4 A0A182XK54 A0A182TYB1 A0A182VN55 A0A182TJV7 A0A1Q3EW18 A0A1Q3EW06 A0A182JZF4 A0A182H1V1 A0A182GLG7 A0A1A9WR38 Q17CA8 A0A1S4F873 A0A182R829 B4PPM7 A0A1B0AGG3 A0A182PMX8 A0A182IV77 A0A1A9V391 A0A1A9XFG3 Q9VAJ1 B3P5F4 A0A1B0BAP1 A0A2M4CNX8 A0A182NKW6 B4HZC2 B4R065 A0A182F8L8 B4KND6 W5JSJ6 B4LMA5 A0A067RAS5 B4J6N9 A0A336LTZ0 T2MD05 A0A1W4UPK9 A7SNA1 A0A0R3NXR9 K1R247 B5DIE4 B4G9A8 A0A182KX05 A0A3B0K6E9 A0A0M5J6X5 R7UQL7 B3MVT7 W4Y6G0 A0A2P2I0B3 E9G738 B0W3E5 A0A0P5K6P8 A0A0P6D2U0 A0A0N8BYD1 A0A0P5F9B2 A0A0P5BBE8 A0A0P5MKM6
A0A194PQ56 A0A0L7KRA2 A0A1I8PTP0 A0A1I8PTS8 A0A1I8MCN6 W8CBD0 A0A182MHJ2 A0A182VSA9 A0A182HJT6 Q7QDI4 A0A182XK54 A0A182TYB1 A0A182VN55 A0A182TJV7 A0A1Q3EW18 A0A1Q3EW06 A0A182JZF4 A0A182H1V1 A0A182GLG7 A0A1A9WR38 Q17CA8 A0A1S4F873 A0A182R829 B4PPM7 A0A1B0AGG3 A0A182PMX8 A0A182IV77 A0A1A9V391 A0A1A9XFG3 Q9VAJ1 B3P5F4 A0A1B0BAP1 A0A2M4CNX8 A0A182NKW6 B4HZC2 B4R065 A0A182F8L8 B4KND6 W5JSJ6 B4LMA5 A0A067RAS5 B4J6N9 A0A336LTZ0 T2MD05 A0A1W4UPK9 A7SNA1 A0A0R3NXR9 K1R247 B5DIE4 B4G9A8 A0A182KX05 A0A3B0K6E9 A0A0M5J6X5 R7UQL7 B3MVT7 W4Y6G0 A0A2P2I0B3 E9G738 B0W3E5 A0A0P5K6P8 A0A0P6D2U0 A0A0N8BYD1 A0A0P5F9B2 A0A0P5BBE8 A0A0P5MKM6
Pubmed
19121390
23622113
22118469
26354079
26227816
25315136
+ More
24495485 12364791 14747013 17210077 26483478 17510324 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 24845553 24065732 17615350 15632085 22992520 20966253 23254933 21292972
24495485 12364791 14747013 17210077 26483478 17510324 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 24845553 24065732 17615350 15632085 22992520 20966253 23254933 21292972
EMBL
BABH01013924
BABH01013925
GAIX01008599
JAA83961.1
ODYU01006382
SOQ48183.1
+ More
RSAL01000163 RVE45375.1 AGBW02014132 OWR42220.1 KQ461154 KPJ08507.1 KQ459595 KPI95576.1 JTDY01006991 KOB65544.1 GAMC01004931 JAC01625.1 AXCM01001279 APCN01002209 AAAB01008851 EAA07299.5 GFDL01015541 JAV19504.1 GFDL01015548 JAV19497.1 JXUM01023258 KQ560656 KXJ81410.1 JXUM01071935 KQ562693 KXJ75324.1 CH477310 EAT43967.1 CM000160 EDW98277.1 AE014297 AY122183 AAF56915.1 AAM52695.1 CH954182 EDV53204.1 JXJN01011087 GGFL01002858 MBW67036.1 CH480819 EDW53379.1 CM000364 EDX14871.1 CH933808 EDW09989.1 ADMH02000563 ETN65880.1 CH940648 EDW60983.1 KK852817 KDR15738.1 CH916367 EDW00942.1 UFQT01000109 SSX20143.1 HAAD01003777 CDG70009.1 DS469718 EDO34797.1 CH379060 KRT04159.1 JH817402 EKC37559.1 EDY70085.1 CH479180 EDW28938.1 OUUW01000004 SPP79048.1 CP012524 ALC40848.1 AMQN01001095 KB299137 ELU08403.1 CH902625 EDV35082.1 AAGJ04101735 AAGJ04101736 AAGJ04101737 AAGJ04101738 AAGJ04101739 AAGJ04101740 IACF01001799 LAB67478.1 GL732534 EFX84401.1 DS231831 EDS31288.1 GDIQ01188335 GDIQ01188334 JAK63391.1 GDIQ01085978 JAN08759.1 GDIQ01132811 JAL18915.1 GDIQ01258167 JAJ93557.1 GDIP01186823 LRGB01002019 JAJ36579.1 KZS09603.1 GDIQ01157948 JAK93777.1
RSAL01000163 RVE45375.1 AGBW02014132 OWR42220.1 KQ461154 KPJ08507.1 KQ459595 KPI95576.1 JTDY01006991 KOB65544.1 GAMC01004931 JAC01625.1 AXCM01001279 APCN01002209 AAAB01008851 EAA07299.5 GFDL01015541 JAV19504.1 GFDL01015548 JAV19497.1 JXUM01023258 KQ560656 KXJ81410.1 JXUM01071935 KQ562693 KXJ75324.1 CH477310 EAT43967.1 CM000160 EDW98277.1 AE014297 AY122183 AAF56915.1 AAM52695.1 CH954182 EDV53204.1 JXJN01011087 GGFL01002858 MBW67036.1 CH480819 EDW53379.1 CM000364 EDX14871.1 CH933808 EDW09989.1 ADMH02000563 ETN65880.1 CH940648 EDW60983.1 KK852817 KDR15738.1 CH916367 EDW00942.1 UFQT01000109 SSX20143.1 HAAD01003777 CDG70009.1 DS469718 EDO34797.1 CH379060 KRT04159.1 JH817402 EKC37559.1 EDY70085.1 CH479180 EDW28938.1 OUUW01000004 SPP79048.1 CP012524 ALC40848.1 AMQN01001095 KB299137 ELU08403.1 CH902625 EDV35082.1 AAGJ04101735 AAGJ04101736 AAGJ04101737 AAGJ04101738 AAGJ04101739 AAGJ04101740 IACF01001799 LAB67478.1 GL732534 EFX84401.1 DS231831 EDS31288.1 GDIQ01188335 GDIQ01188334 JAK63391.1 GDIQ01085978 JAN08759.1 GDIQ01132811 JAL18915.1 GDIQ01258167 JAJ93557.1 GDIP01186823 LRGB01002019 JAJ36579.1 KZS09603.1 GDIQ01157948 JAK93777.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000095300 UP000095301 UP000075883 UP000075920 UP000075840 UP000007062 UP000076407 UP000075902 UP000075903 UP000075881 UP000069940 UP000249989 UP000091820 UP000008820 UP000075900 UP000002282 UP000092445 UP000075885 UP000075880 UP000078200 UP000092443 UP000000803 UP000008711 UP000092460 UP000075884 UP000001292 UP000000304 UP000069272 UP000009192 UP000000673 UP000008792 UP000027135 UP000001070 UP000192221 UP000001593 UP000001819 UP000005408 UP000008744 UP000075882 UP000268350 UP000092553 UP000014760 UP000007801 UP000007110 UP000000305 UP000002320 UP000076858
UP000095300 UP000095301 UP000075883 UP000075920 UP000075840 UP000007062 UP000076407 UP000075902 UP000075903 UP000075881 UP000069940 UP000249989 UP000091820 UP000008820 UP000075900 UP000002282 UP000092445 UP000075885 UP000075880 UP000078200 UP000092443 UP000000803 UP000008711 UP000092460 UP000075884 UP000001292 UP000000304 UP000069272 UP000009192 UP000000673 UP000008792 UP000027135 UP000001070 UP000192221 UP000001593 UP000001819 UP000005408 UP000008744 UP000075882 UP000268350 UP000092553 UP000014760 UP000007801 UP000007110 UP000000305 UP000002320 UP000076858
PRIDE
Pfam
Interpro
IPR016024
ARM-type_fold
+ More
IPR032682 Cnd1_C
IPR026971 CND1/NCAPD3
IPR007673 Condensin_cplx_su1
IPR015797 NUDIX_hydrolase-like_dom_sf
IPR011989 ARM-like
IPR000086 NUDIX_hydrolase_dom
IPR024324 Condensin_cplx_su1_N
IPR027417 P-loop_NTPase
IPR031314 DNK_dom
IPR001313 Pumilio_RNA-bd_rpt
IPR026003 Cohesin_HEAT
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR000048 IQ_motif_EF-hand-BS
IPR005559 CG-1_dom
IPR002909 IPT_dom
IPR014756 Ig_E-set
IPR013783 Ig-like_fold
IPR032682 Cnd1_C
IPR026971 CND1/NCAPD3
IPR007673 Condensin_cplx_su1
IPR015797 NUDIX_hydrolase-like_dom_sf
IPR011989 ARM-like
IPR000086 NUDIX_hydrolase_dom
IPR024324 Condensin_cplx_su1_N
IPR027417 P-loop_NTPase
IPR031314 DNK_dom
IPR001313 Pumilio_RNA-bd_rpt
IPR026003 Cohesin_HEAT
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR000048 IQ_motif_EF-hand-BS
IPR005559 CG-1_dom
IPR002909 IPT_dom
IPR014756 Ig_E-set
IPR013783 Ig-like_fold
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9J7J2
S4NYG7
A0A2H1W582
A0A3S2NV77
A0A212EL47
A0A194QSP9
+ More
A0A194PQ56 A0A0L7KRA2 A0A1I8PTP0 A0A1I8PTS8 A0A1I8MCN6 W8CBD0 A0A182MHJ2 A0A182VSA9 A0A182HJT6 Q7QDI4 A0A182XK54 A0A182TYB1 A0A182VN55 A0A182TJV7 A0A1Q3EW18 A0A1Q3EW06 A0A182JZF4 A0A182H1V1 A0A182GLG7 A0A1A9WR38 Q17CA8 A0A1S4F873 A0A182R829 B4PPM7 A0A1B0AGG3 A0A182PMX8 A0A182IV77 A0A1A9V391 A0A1A9XFG3 Q9VAJ1 B3P5F4 A0A1B0BAP1 A0A2M4CNX8 A0A182NKW6 B4HZC2 B4R065 A0A182F8L8 B4KND6 W5JSJ6 B4LMA5 A0A067RAS5 B4J6N9 A0A336LTZ0 T2MD05 A0A1W4UPK9 A7SNA1 A0A0R3NXR9 K1R247 B5DIE4 B4G9A8 A0A182KX05 A0A3B0K6E9 A0A0M5J6X5 R7UQL7 B3MVT7 W4Y6G0 A0A2P2I0B3 E9G738 B0W3E5 A0A0P5K6P8 A0A0P6D2U0 A0A0N8BYD1 A0A0P5F9B2 A0A0P5BBE8 A0A0P5MKM6
A0A194PQ56 A0A0L7KRA2 A0A1I8PTP0 A0A1I8PTS8 A0A1I8MCN6 W8CBD0 A0A182MHJ2 A0A182VSA9 A0A182HJT6 Q7QDI4 A0A182XK54 A0A182TYB1 A0A182VN55 A0A182TJV7 A0A1Q3EW18 A0A1Q3EW06 A0A182JZF4 A0A182H1V1 A0A182GLG7 A0A1A9WR38 Q17CA8 A0A1S4F873 A0A182R829 B4PPM7 A0A1B0AGG3 A0A182PMX8 A0A182IV77 A0A1A9V391 A0A1A9XFG3 Q9VAJ1 B3P5F4 A0A1B0BAP1 A0A2M4CNX8 A0A182NKW6 B4HZC2 B4R065 A0A182F8L8 B4KND6 W5JSJ6 B4LMA5 A0A067RAS5 B4J6N9 A0A336LTZ0 T2MD05 A0A1W4UPK9 A7SNA1 A0A0R3NXR9 K1R247 B5DIE4 B4G9A8 A0A182KX05 A0A3B0K6E9 A0A0M5J6X5 R7UQL7 B3MVT7 W4Y6G0 A0A2P2I0B3 E9G738 B0W3E5 A0A0P5K6P8 A0A0P6D2U0 A0A0N8BYD1 A0A0P5F9B2 A0A0P5BBE8 A0A0P5MKM6
Ontologies
GO
GO:0007076
GO:0005634
GO:0016787
GO:0051301
GO:0007049
GO:0030261
GO:0010032
GO:0000799
GO:0042393
GO:0003682
GO:0051304
GO:0000779
GO:0000785
GO:0000796
GO:0005737
GO:0000070
GO:0007419
GO:0003723
GO:0003677
GO:0005488
GO:0016021
GO:0020037
GO:0016491
GO:0007165
GO:0015031
GO:0016020
GO:0016742
GO:0003824
PANTHER
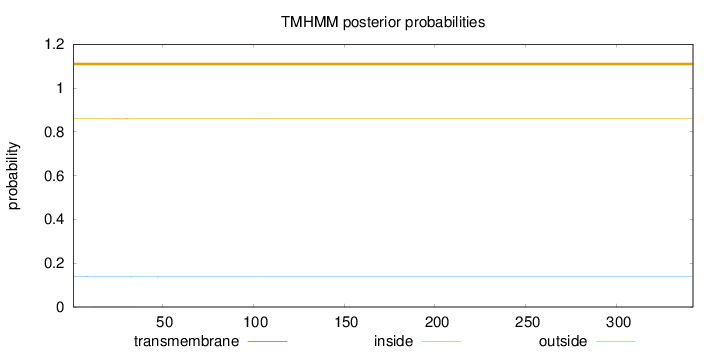
Topology
Length:
342
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01496
Exp number, first 60 AAs:
0.01433
Total prob of N-in:
0.13850
outside
1 - 342
Population Genetic Test Statistics
Pi
177.219062
Theta
169.352719
Tajima's D
0.415397
CLR
0.039216
CSRT
0.489825508724564
Interpretation
Uncertain
