Pre Gene Modal
BGIBMGA005328
Annotation
translocon-associated_protein_gamma_isoform_1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.513
Sequence
CDS
ATGTCTGGAAAGAACAACAAAGCTTTTACAAAAGAAGAAGAGTTATTGTTGCAAGATTTTAGCCGGAATGTGTCCACAAAATCGTCTGCTCTCTTTTACGGGAACGCTTTTATTGTTTCAGCAATTCCAATTTGGTTGTTCTGGAGGGTACATTCATTGGAAATAAGCACCGCAATTATTTGGTTTATCCTAGTAACGGCAGCTAGCACCTGGTTGCTGGCTCTTGCCTACCGTAACACTAAGTTCCAGTTGAAGCACCGTGTTGCTGTGCGCAGGGAGGATGCTGTTGCCCGTGAAATGTCCAGGAAGCTTGCTGATGATAAAAAAATGAGCAGAAAAGAAAAGGACGAAAGAATCTTGTGGAAAAAGAATGAGGTGGCAGATTATGAAGCTACCACATACTCAATTTTCTACAATAACGCACTTTTCCTGACTATTGTCATCTTGAGCAGTTTCTACATTCTGCGTACATTCACACCTACAGTTAATTACATTGTGTCTCTCACGGCTGCATCTGGACTTCTTGCTCTACTCTCCACAGGAACCAAGTGA
Protein
MSGKNNKAFTKEEELLLQDFSRNVSTKSSALFYGNAFIVSAIPIWLFWRVHSLEISTAIIWFILVTAASTWLLALAYRNTKFQLKHRVAVRREDAVAREMSRKLADDKKMSRKEKDERILWKKNEVADYEATTYSIFYNNALFLTIVILSSFYILRTFTPTVNYIVSLTAASGLLALLSTGTK
Summary
Uniprot
Q2F5K1
H9J736
Q2F5K0
A0A3G1T1G7
A0A1E1W5V6
A0A194QY53
+ More
I4DIY8 I4DJA7 S4P4H2 A0A3S2NEH2 A0A212EL61 A0A2H1W5S0 A0A2A4IT47 A0A0L7LAY1 A0A067RIR7 A0A2J7PBJ7 A0A2P8XN60 A0A0M9A255 A0A154PQN7 A0A2A3EN45 V9IF99 U3KR34 A0A0L7RJ97 A0A195CL98 E2A0K0 A0A195D9X3 F4WZ33 A0A195BQT1 A0A158NDN1 A0A151XEK6 A0A0L7QJX6 V5G6J0 E9IYV1 A0A3L8DZN5 A0A1D2MVJ5 A0A067RKR5 N6TG74 E2C5D7 A0A1B6EM72 A0A226E3Z6 D7EL46 A0A0C9PSB0 K7J1T1 A0A1B6K155 A0A1B6LN78 E0VK54 A0A232F6S2 U5ETN8 A0A069DNW4 R4G8L4 A0A1B6DWE4 A0A023EIJ2 R4WIR1 A0A0A9WYI6 A0A224XZ73 A0A023FAI7 A0A1S3IS39 A0A1D1YDY7 Q1HRN7 A0A1B6MQ12 A0A1L8DUR7 A0A1W4WVP4 A0A182RVS0 A0A182YM22 B4MZD6 A0A1Y1MJE8 A0A026VZ11 A0A182T648 A0A1Q3F9W0 A0A0L0CHP8 A0A1B0DDP8 A0A182KGD3 A0A0K8W808 A0A034WEV7 A0A0C9Q6B6 A0A293N222 B0WXG2 A0A195F4Y4 A0A084VMP8 A0A182LVJ0 A0A182P2U2 A0A2I9LPV3 A0A0A1XPR7 W8B847 Q7Q876 A0A1E1XS16 A0A1E1WYV3 A0A182VTK9 A0A1S4GZ67 A0A182X651 A0A182KWL4 A0A182IB73 A0A2R5LJW2 A0A023FX36 A0A023G1U9 A0A0K8TPY3 T1P7N5 A0A1I8NC39 A0A1L8EFC8 B3MPA7
I4DIY8 I4DJA7 S4P4H2 A0A3S2NEH2 A0A212EL61 A0A2H1W5S0 A0A2A4IT47 A0A0L7LAY1 A0A067RIR7 A0A2J7PBJ7 A0A2P8XN60 A0A0M9A255 A0A154PQN7 A0A2A3EN45 V9IF99 U3KR34 A0A0L7RJ97 A0A195CL98 E2A0K0 A0A195D9X3 F4WZ33 A0A195BQT1 A0A158NDN1 A0A151XEK6 A0A0L7QJX6 V5G6J0 E9IYV1 A0A3L8DZN5 A0A1D2MVJ5 A0A067RKR5 N6TG74 E2C5D7 A0A1B6EM72 A0A226E3Z6 D7EL46 A0A0C9PSB0 K7J1T1 A0A1B6K155 A0A1B6LN78 E0VK54 A0A232F6S2 U5ETN8 A0A069DNW4 R4G8L4 A0A1B6DWE4 A0A023EIJ2 R4WIR1 A0A0A9WYI6 A0A224XZ73 A0A023FAI7 A0A1S3IS39 A0A1D1YDY7 Q1HRN7 A0A1B6MQ12 A0A1L8DUR7 A0A1W4WVP4 A0A182RVS0 A0A182YM22 B4MZD6 A0A1Y1MJE8 A0A026VZ11 A0A182T648 A0A1Q3F9W0 A0A0L0CHP8 A0A1B0DDP8 A0A182KGD3 A0A0K8W808 A0A034WEV7 A0A0C9Q6B6 A0A293N222 B0WXG2 A0A195F4Y4 A0A084VMP8 A0A182LVJ0 A0A182P2U2 A0A2I9LPV3 A0A0A1XPR7 W8B847 Q7Q876 A0A1E1XS16 A0A1E1WYV3 A0A182VTK9 A0A1S4GZ67 A0A182X651 A0A182KWL4 A0A182IB73 A0A2R5LJW2 A0A023FX36 A0A023G1U9 A0A0K8TPY3 T1P7N5 A0A1I8NC39 A0A1L8EFC8 B3MPA7
Pubmed
19121390
26354079
22651552
23622113
22118469
26227816
+ More
24845553 29403074 20798317 21719571 21347285 21282665 30249741 27289101 23537049 18362917 19820115 20075255 20566863 28648823 26334808 24945155 23691247 25401762 26823975 25474469 17204158 17510324 25244985 17994087 28004739 24508170 26108605 25348373 24438588 29248469 25830018 24495485 12364791 29209593 28503490 20966253 26369729 25315136
24845553 29403074 20798317 21719571 21347285 21282665 30249741 27289101 23537049 18362917 19820115 20075255 20566863 28648823 26334808 24945155 23691247 25401762 26823975 25474469 17204158 17510324 25244985 17994087 28004739 24508170 26108605 25348373 24438588 29248469 25830018 24495485 12364791 29209593 28503490 20966253 26369729 25315136
EMBL
DQ311422
ABD36366.1
BABH01013922
DQ311423
ABD36367.1
MG992374
+ More
AXY94812.1 GDQN01008725 JAT82329.1 KQ461154 KPJ08506.1 AK401256 KQ459595 BAM17878.1 KPI95575.1 AK401375 BAM17997.1 GAIX01005964 JAA86596.1 RSAL01000163 RVE45374.1 AGBW02014132 OWR42219.1 ODYU01006382 SOQ48182.1 NWSH01007489 PCG62945.1 JTDY01001885 KOB72632.1 KK852452 KDR23667.1 NEVH01027081 PNF13716.1 PYGN01001675 PSN33432.1 KQ435776 KOX74886.1 KQ435050 KZC14235.1 KZ288215 PBC32682.1 JR043246 AEY59745.1 KQ414580 KOC71047.1 KQ977600 KYN01490.1 GL435626 EFN72903.1 KQ981082 KYN09720.1 GL888465 EGI60536.1 KQ976424 KYM88319.1 ADTU01012727 KQ982254 KYQ58787.1 KQ415027 KOC58928.1 GALX01002766 JAB65700.1 GL767051 EFZ14259.1 QOIP01000002 RLU25693.1 LJIJ01000500 ODM96824.1 KK852413 KDR24422.1 APGK01039052 KB740967 KB632399 ENN76773.1 ERL94740.1 GL452770 EFN76870.1 GECZ01030742 GECZ01006095 JAS39027.1 JAS63674.1 LNIX01000007 OXA51998.1 DS497806 EFA11879.1 GBYB01004243 JAG74010.1 GECU01002548 JAT05159.1 GEBQ01014824 JAT25153.1 DS235239 EEB13760.1 NNAY01000817 OXU26345.1 GANO01002693 JAB57178.1 GBGD01003131 JAC85758.1 GAHY01000767 JAA76743.1 GEDC01007316 JAS29982.1 GAPW01004833 JAC08765.1 AK417350 BAN20565.1 GBHO01030077 GBRD01012544 GDHC01019300 JAG13527.1 JAG53280.1 JAP99328.1 GFTR01003027 JAW13399.1 GBBI01000683 JAC18029.1 GDJX01015088 JAT52848.1 DQ440057 CH477441 ABF18090.1 EAT40851.1 EJY57689.1 GEBQ01001940 JAT38037.1 GFDF01003916 JAV10168.1 CH963913 EDW77409.1 GEZM01032222 GEZM01032221 JAV84760.1 KK107578 EZA48686.1 GFDL01010719 JAV24326.1 JRES01000384 KNC31742.1 AJVK01032139 GDHF01005068 JAI47246.1 GAKP01004841 JAC54111.1 GBYB01009658 JAG79425.1 GFWV01022437 MAA47164.1 DS232164 EDS36492.1 KQ981820 KYN35139.1 ATLV01014616 KE524975 KFB39242.1 AXCM01000488 GFWZ01000425 MBW20415.1 GBXI01001447 JAD12845.1 GAMC01011753 JAB94802.1 AAAB01008944 EAA10212.2 GFAA01001356 JAU02079.1 GFAC01007026 JAT92162.1 APCN01000644 GGLE01005704 MBY09830.1 GBBL01002012 JAC25308.1 GBBM01007247 JAC28171.1 GDAI01001412 JAI16191.1 KA644484 AFP59113.1 GFDG01001372 JAV17427.1 CH902620 EDV32226.1
AXY94812.1 GDQN01008725 JAT82329.1 KQ461154 KPJ08506.1 AK401256 KQ459595 BAM17878.1 KPI95575.1 AK401375 BAM17997.1 GAIX01005964 JAA86596.1 RSAL01000163 RVE45374.1 AGBW02014132 OWR42219.1 ODYU01006382 SOQ48182.1 NWSH01007489 PCG62945.1 JTDY01001885 KOB72632.1 KK852452 KDR23667.1 NEVH01027081 PNF13716.1 PYGN01001675 PSN33432.1 KQ435776 KOX74886.1 KQ435050 KZC14235.1 KZ288215 PBC32682.1 JR043246 AEY59745.1 KQ414580 KOC71047.1 KQ977600 KYN01490.1 GL435626 EFN72903.1 KQ981082 KYN09720.1 GL888465 EGI60536.1 KQ976424 KYM88319.1 ADTU01012727 KQ982254 KYQ58787.1 KQ415027 KOC58928.1 GALX01002766 JAB65700.1 GL767051 EFZ14259.1 QOIP01000002 RLU25693.1 LJIJ01000500 ODM96824.1 KK852413 KDR24422.1 APGK01039052 KB740967 KB632399 ENN76773.1 ERL94740.1 GL452770 EFN76870.1 GECZ01030742 GECZ01006095 JAS39027.1 JAS63674.1 LNIX01000007 OXA51998.1 DS497806 EFA11879.1 GBYB01004243 JAG74010.1 GECU01002548 JAT05159.1 GEBQ01014824 JAT25153.1 DS235239 EEB13760.1 NNAY01000817 OXU26345.1 GANO01002693 JAB57178.1 GBGD01003131 JAC85758.1 GAHY01000767 JAA76743.1 GEDC01007316 JAS29982.1 GAPW01004833 JAC08765.1 AK417350 BAN20565.1 GBHO01030077 GBRD01012544 GDHC01019300 JAG13527.1 JAG53280.1 JAP99328.1 GFTR01003027 JAW13399.1 GBBI01000683 JAC18029.1 GDJX01015088 JAT52848.1 DQ440057 CH477441 ABF18090.1 EAT40851.1 EJY57689.1 GEBQ01001940 JAT38037.1 GFDF01003916 JAV10168.1 CH963913 EDW77409.1 GEZM01032222 GEZM01032221 JAV84760.1 KK107578 EZA48686.1 GFDL01010719 JAV24326.1 JRES01000384 KNC31742.1 AJVK01032139 GDHF01005068 JAI47246.1 GAKP01004841 JAC54111.1 GBYB01009658 JAG79425.1 GFWV01022437 MAA47164.1 DS232164 EDS36492.1 KQ981820 KYN35139.1 ATLV01014616 KE524975 KFB39242.1 AXCM01000488 GFWZ01000425 MBW20415.1 GBXI01001447 JAD12845.1 GAMC01011753 JAB94802.1 AAAB01008944 EAA10212.2 GFAA01001356 JAU02079.1 GFAC01007026 JAT92162.1 APCN01000644 GGLE01005704 MBY09830.1 GBBL01002012 JAC25308.1 GBBM01007247 JAC28171.1 GDAI01001412 JAI16191.1 KA644484 AFP59113.1 GFDG01001372 JAV17427.1 CH902620 EDV32226.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000007151
UP000218220
+ More
UP000037510 UP000027135 UP000235965 UP000245037 UP000053105 UP000076502 UP000242457 UP000005203 UP000053825 UP000078542 UP000000311 UP000078492 UP000007755 UP000078540 UP000005205 UP000075809 UP000279307 UP000094527 UP000019118 UP000030742 UP000008237 UP000198287 UP000007266 UP000002358 UP000009046 UP000215335 UP000085678 UP000008820 UP000192223 UP000075900 UP000076408 UP000007798 UP000053097 UP000075901 UP000037069 UP000092462 UP000075881 UP000002320 UP000078541 UP000030765 UP000075883 UP000075885 UP000007062 UP000075920 UP000076407 UP000075882 UP000075840 UP000095301 UP000007801
UP000037510 UP000027135 UP000235965 UP000245037 UP000053105 UP000076502 UP000242457 UP000005203 UP000053825 UP000078542 UP000000311 UP000078492 UP000007755 UP000078540 UP000005205 UP000075809 UP000279307 UP000094527 UP000019118 UP000030742 UP000008237 UP000198287 UP000007266 UP000002358 UP000009046 UP000215335 UP000085678 UP000008820 UP000192223 UP000075900 UP000076408 UP000007798 UP000053097 UP000075901 UP000037069 UP000092462 UP000075881 UP000002320 UP000078541 UP000030765 UP000075883 UP000075885 UP000007062 UP000075920 UP000076407 UP000075882 UP000075840 UP000095301 UP000007801
Interpro
SUPFAM
SSF103473
SSF103473
Gene 3D
ProteinModelPortal
Q2F5K1
H9J736
Q2F5K0
A0A3G1T1G7
A0A1E1W5V6
A0A194QY53
+ More
I4DIY8 I4DJA7 S4P4H2 A0A3S2NEH2 A0A212EL61 A0A2H1W5S0 A0A2A4IT47 A0A0L7LAY1 A0A067RIR7 A0A2J7PBJ7 A0A2P8XN60 A0A0M9A255 A0A154PQN7 A0A2A3EN45 V9IF99 U3KR34 A0A0L7RJ97 A0A195CL98 E2A0K0 A0A195D9X3 F4WZ33 A0A195BQT1 A0A158NDN1 A0A151XEK6 A0A0L7QJX6 V5G6J0 E9IYV1 A0A3L8DZN5 A0A1D2MVJ5 A0A067RKR5 N6TG74 E2C5D7 A0A1B6EM72 A0A226E3Z6 D7EL46 A0A0C9PSB0 K7J1T1 A0A1B6K155 A0A1B6LN78 E0VK54 A0A232F6S2 U5ETN8 A0A069DNW4 R4G8L4 A0A1B6DWE4 A0A023EIJ2 R4WIR1 A0A0A9WYI6 A0A224XZ73 A0A023FAI7 A0A1S3IS39 A0A1D1YDY7 Q1HRN7 A0A1B6MQ12 A0A1L8DUR7 A0A1W4WVP4 A0A182RVS0 A0A182YM22 B4MZD6 A0A1Y1MJE8 A0A026VZ11 A0A182T648 A0A1Q3F9W0 A0A0L0CHP8 A0A1B0DDP8 A0A182KGD3 A0A0K8W808 A0A034WEV7 A0A0C9Q6B6 A0A293N222 B0WXG2 A0A195F4Y4 A0A084VMP8 A0A182LVJ0 A0A182P2U2 A0A2I9LPV3 A0A0A1XPR7 W8B847 Q7Q876 A0A1E1XS16 A0A1E1WYV3 A0A182VTK9 A0A1S4GZ67 A0A182X651 A0A182KWL4 A0A182IB73 A0A2R5LJW2 A0A023FX36 A0A023G1U9 A0A0K8TPY3 T1P7N5 A0A1I8NC39 A0A1L8EFC8 B3MPA7
I4DIY8 I4DJA7 S4P4H2 A0A3S2NEH2 A0A212EL61 A0A2H1W5S0 A0A2A4IT47 A0A0L7LAY1 A0A067RIR7 A0A2J7PBJ7 A0A2P8XN60 A0A0M9A255 A0A154PQN7 A0A2A3EN45 V9IF99 U3KR34 A0A0L7RJ97 A0A195CL98 E2A0K0 A0A195D9X3 F4WZ33 A0A195BQT1 A0A158NDN1 A0A151XEK6 A0A0L7QJX6 V5G6J0 E9IYV1 A0A3L8DZN5 A0A1D2MVJ5 A0A067RKR5 N6TG74 E2C5D7 A0A1B6EM72 A0A226E3Z6 D7EL46 A0A0C9PSB0 K7J1T1 A0A1B6K155 A0A1B6LN78 E0VK54 A0A232F6S2 U5ETN8 A0A069DNW4 R4G8L4 A0A1B6DWE4 A0A023EIJ2 R4WIR1 A0A0A9WYI6 A0A224XZ73 A0A023FAI7 A0A1S3IS39 A0A1D1YDY7 Q1HRN7 A0A1B6MQ12 A0A1L8DUR7 A0A1W4WVP4 A0A182RVS0 A0A182YM22 B4MZD6 A0A1Y1MJE8 A0A026VZ11 A0A182T648 A0A1Q3F9W0 A0A0L0CHP8 A0A1B0DDP8 A0A182KGD3 A0A0K8W808 A0A034WEV7 A0A0C9Q6B6 A0A293N222 B0WXG2 A0A195F4Y4 A0A084VMP8 A0A182LVJ0 A0A182P2U2 A0A2I9LPV3 A0A0A1XPR7 W8B847 Q7Q876 A0A1E1XS16 A0A1E1WYV3 A0A182VTK9 A0A1S4GZ67 A0A182X651 A0A182KWL4 A0A182IB73 A0A2R5LJW2 A0A023FX36 A0A023G1U9 A0A0K8TPY3 T1P7N5 A0A1I8NC39 A0A1L8EFC8 B3MPA7
Ontologies
GO
PANTHER
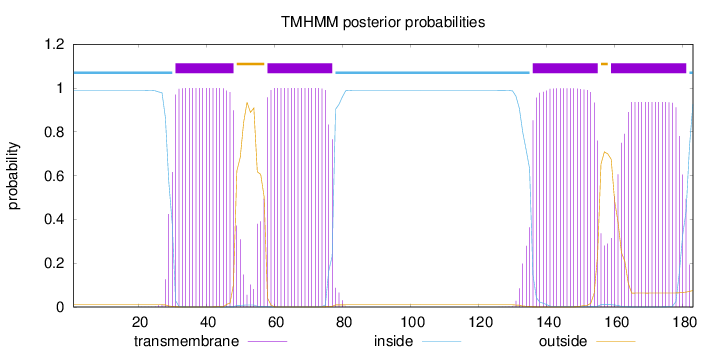
Topology
Length:
183
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
81.3317699999999
Exp number, first 60 AAs:
24.2962
Total prob of N-in:
0.98949
POSSIBLE N-term signal
sequence
inside
1 - 30
TMhelix
31 - 48
outside
49 - 57
TMhelix
58 - 77
inside
78 - 135
TMhelix
136 - 155
outside
156 - 158
TMhelix
159 - 181
inside
182 - 183
Population Genetic Test Statistics
Pi
38.33821
Theta
30.305897
Tajima's D
-0.517882
CLR
150.902374
CSRT
0.23958802059897
Interpretation
Uncertain
