Gene
KWMTBOMO04583
Pre Gene Modal
BGIBMGA005482
Annotation
PREDICTED:_uncharacterized_protein_LOC106141078_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.167
Sequence
CDS
ATGCTGGCTAGTGCACTTGCAGCACGCTTTGGCGACCGCCTCGAGCCGCGGATGCCTGCCGCATACGCACCGCGGAATGTACAAACCGAACACAACGTAGCAGAGACTGACTACGAAAATGCGTCATATCACGAATATGACGAAGAAAATGCCTCAATCGAAGAAAACAATACAAAACCAGAAACAGAACCATCACCCCGTCGACCCACCCGCACAGAAGCTCATCGGCTTGAAATGAGTACAGAATATTTCGTAATCCCGAAACGCACAACTACGACATCGACGATTTCCACTTCGACGTCATCGACTGAGCCGTCGCCATCTCAGTCACCAAACAACAATACTAACACTCCTGCAACTATTGCAAAAACAGCGGGAGTGCTAAGAACGGAACCATGGGCTGTCCCAATTTTAGCATTAGCTAGTGTGATTATGGTGATTCTTGGTGCTTTCGAGGCATTTGTAATTTGGGGGGCCAGCCGGAAAGCTCCGAGTCAACGTCATTTACTTCTAGGACAGACATTGCTTTTTGGGCTATTCTTATGTGCCGCCACGGCAGCACTATTTGCCACAACACCAACTGCATTTACCTGTGGCGCAGTTCGATTCGGAACCGGCGTAGCCTATGTAATAGTGTTTGCATCCCTTCTAGTGAAGTGCGTTTTCCTACTGAGTTTGAATGGTGGAATTTATCTGCCGGCAGCGTATCAAGGACTGCTTTTATTTTTTGCTGTAATGATACAAGTAGCAATTGGTGCTCAGTGGCTCGGTGGTTCTCCTCCGAGAGTCATTAACGGAGGAGGTCGGTGTGATTGTCAACTATCAGATCTTCTTATGTCACTGTGTTACGCAGCTTTTTTAATAGCTGTGGTGTGTGGTGTAGCATTACGATCTCGAGGAATTCGTAATAACTATCGAGAAGCAACGCATATAACTGCTGCAGGGGGCGCTACCGCTGCAGTATGGGTGTGTTGGATAGGAGCCTCGCTAGGTGCTCCTGAGCGCCATCGCGATGCAAGCGTGGCAGCTGGTTTACTTGCAACATGCGCAGTAGTCTTTGCCCTTATGTTCGGACCAAAAGGTCGACGTTTAGCGGCCCTGGGACGAGAAGGTCGATGGGACGCAGATCGTGAGGAAGGGCTAAGTTCCCTTGGAGCCGGTGGATCTGGGTACTCTCCTTCATTCTTTCATTTCAAACCAGTCAAGTATGGAATGGTATCAGCGACAGCTGCAACTTCCGTTCCTCCACCAGCACTCGATCGCAAGGAACAGCCTGCTCAGCCAGCCGATCCGTATGGGGCCATGTTCTCAAATTCGCAGCACCCGCACTCACGCGTTTTGTATTCTCCGCCGCACTATTCTTTGCACCACCTAATGCACTATAACTACAACTATCCACCTTTTCCTTATATGCTGCCACCAGGCGTGATGGTGCGTCCGGAGGAGGGCAACGTGTACACGAGCGTAGAACCCACCTTCAGCAGCAACCCCAACGTATACTTCCAACGCACCGAGCCGTTGCACGTCGGCATGATGTACTGA
Protein
MLASALAARFGDRLEPRMPAAYAPRNVQTEHNVAETDYENASYHEYDEENASIEENNTKPETEPSPRRPTRTEAHRLEMSTEYFVIPKRTTTTSTISTSTSSTEPSPSQSPNNNTNTPATIAKTAGVLRTEPWAVPILALASVIMVILGAFEAFVIWGASRKAPSQRHLLLGQTLLFGLFLCAATAALFATTPTAFTCGAVRFGTGVAYVIVFASLLVKCVFLLSLNGGIYLPAAYQGLLLFFAVMIQVAIGAQWLGGSPPRVINGGGRCDCQLSDLLMSLCYAAFLIAVVCGVALRSRGIRNNYREATHITAAGGATAAVWVCWIGASLGAPERHRDASVAAGLLATCAVVFALMFGPKGRRLAALGREGRWDADREEGLSSLGAGGSGYSPSFFHFKPVKYGMVSATAATSVPPPALDRKEQPAQPADPYGAMFSNSQHPHSRVLYSPPHYSLHHLMHYNYNYPPFPYMLPPGVMVRPEEGNVYTSVEPTFSSNPNVYFQRTEPLHVGMMY
Summary
Uniprot
H9J7J0
A0A3S2L4C5
A0A2H1W525
A0A194PS45
A0A212EL71
A0A194QU36
+ More
I4DMA5 D6WE24 A0A2J7RBE3 A0A2J7RBE7 A0A1Y1MUP5 A0A1I8N4A0 A0A1I8N484 A0A1I8N4A1 A0A1I8Q7S6 A0A1I8Q7S3 A0A1W4X0S3 A0A0J9S0Z4 M9PIJ4 A0A0Q9WKP1 A0A0Q5U5X1 A0A1W4UE79 A0A0Q9XE83 A0A182JLV2 A0A1J1J762 A0A0P8XT72 B4LD89 A0A0R1DYN0 B4KUW8 A0A3B0J3E4 A0A0R3P3X3 A0A1W4WZR5 A0A336MIP6 A0A336L767 A0A0A9YWZ7 A0A0Q9WSK3 A0A0Q9XMS6 B0WM52 A0A0Q9WP33 A0A1B6JFN1 A0A1B6LNM2 Q5TMI1 A0A182KF49 A0A023F3D5 A0A182PWD9 A0A1B6LD82 A0A1B6FZX9 Q17N19 A0A1B6GUY7 B4IYU8 A0A182RQM6 A0A182WX88 A0A182U573 A0A182N2F0 A0A182HN15 A0A2P8ZIZ8 A0A182LNT5 A0A182V7C9 A0A0M5J8R4 A0A0R1DYP2 B4PDI2 T1GGZ3 E0VHU6 A0A232FFR2 K7JHE1 A0A1B0CM31
I4DMA5 D6WE24 A0A2J7RBE3 A0A2J7RBE7 A0A1Y1MUP5 A0A1I8N4A0 A0A1I8N484 A0A1I8N4A1 A0A1I8Q7S6 A0A1I8Q7S3 A0A1W4X0S3 A0A0J9S0Z4 M9PIJ4 A0A0Q9WKP1 A0A0Q5U5X1 A0A1W4UE79 A0A0Q9XE83 A0A182JLV2 A0A1J1J762 A0A0P8XT72 B4LD89 A0A0R1DYN0 B4KUW8 A0A3B0J3E4 A0A0R3P3X3 A0A1W4WZR5 A0A336MIP6 A0A336L767 A0A0A9YWZ7 A0A0Q9WSK3 A0A0Q9XMS6 B0WM52 A0A0Q9WP33 A0A1B6JFN1 A0A1B6LNM2 Q5TMI1 A0A182KF49 A0A023F3D5 A0A182PWD9 A0A1B6LD82 A0A1B6FZX9 Q17N19 A0A1B6GUY7 B4IYU8 A0A182RQM6 A0A182WX88 A0A182U573 A0A182N2F0 A0A182HN15 A0A2P8ZIZ8 A0A182LNT5 A0A182V7C9 A0A0M5J8R4 A0A0R1DYP2 B4PDI2 T1GGZ3 E0VHU6 A0A232FFR2 K7JHE1 A0A1B0CM31
Pubmed
EMBL
BABH01013922
RSAL01000163
RVE45373.1
ODYU01006382
SOQ48181.1
KQ459595
+ More
KPI95574.1 AGBW02014132 OWR42218.1 KQ461154 KPJ08505.1 AK402423 BAM19045.1 KQ971324 EFA01177.1 NEVH01005906 PNF38155.1 PNF38156.1 GEZM01026277 JAV87077.1 CM002912 KMZ01075.1 AE014296 AGB94881.1 CH940647 KRF85240.1 CH954178 KQS44389.1 CH933809 KRG06613.1 CVRI01000074 CRL08223.1 CH902618 KPU77919.1 EDW70996.1 CM000159 KRK02172.1 EDW19374.2 OUUW01000002 SPP76144.1 CH379069 KRT07820.1 UFQS01000631 UFQT01000631 SSX05549.1 SSX25908.1 UFQS01002121 UFQT01002121 SSX13274.1 SSX32711.1 GBHO01011192 GBHO01009519 GBRD01013399 GDHC01006647 JAG32412.1 JAG34085.1 JAG52427.1 JAQ11982.1 KRF85239.1 KRG06612.1 DS231995 EDS30886.1 CH963847 KRF97616.1 GECU01010043 JAS97663.1 GEBQ01014709 JAT25268.1 AAAB01008986 EAL38916.3 GBBI01002971 JAC15741.1 GEBQ01018346 JAT21631.1 GECZ01014156 GECZ01012121 JAS55613.1 JAS57648.1 CH477202 EAT48126.1 GECZ01003511 JAS66258.1 CH916366 EDV96635.1 APCN01001825 PYGN01000041 PSN56476.1 CP012525 ALC45224.1 KRK02171.1 EDW94981.1 CAQQ02043823 DS235172 EEB12952.1 NNAY01000314 OXU29279.1 AJWK01018095 AJWK01018096
KPI95574.1 AGBW02014132 OWR42218.1 KQ461154 KPJ08505.1 AK402423 BAM19045.1 KQ971324 EFA01177.1 NEVH01005906 PNF38155.1 PNF38156.1 GEZM01026277 JAV87077.1 CM002912 KMZ01075.1 AE014296 AGB94881.1 CH940647 KRF85240.1 CH954178 KQS44389.1 CH933809 KRG06613.1 CVRI01000074 CRL08223.1 CH902618 KPU77919.1 EDW70996.1 CM000159 KRK02172.1 EDW19374.2 OUUW01000002 SPP76144.1 CH379069 KRT07820.1 UFQS01000631 UFQT01000631 SSX05549.1 SSX25908.1 UFQS01002121 UFQT01002121 SSX13274.1 SSX32711.1 GBHO01011192 GBHO01009519 GBRD01013399 GDHC01006647 JAG32412.1 JAG34085.1 JAG52427.1 JAQ11982.1 KRF85239.1 KRG06612.1 DS231995 EDS30886.1 CH963847 KRF97616.1 GECU01010043 JAS97663.1 GEBQ01014709 JAT25268.1 AAAB01008986 EAL38916.3 GBBI01002971 JAC15741.1 GEBQ01018346 JAT21631.1 GECZ01014156 GECZ01012121 JAS55613.1 JAS57648.1 CH477202 EAT48126.1 GECZ01003511 JAS66258.1 CH916366 EDV96635.1 APCN01001825 PYGN01000041 PSN56476.1 CP012525 ALC45224.1 KRK02171.1 EDW94981.1 CAQQ02043823 DS235172 EEB12952.1 NNAY01000314 OXU29279.1 AJWK01018095 AJWK01018096
Proteomes
UP000005204
UP000283053
UP000053268
UP000007151
UP000053240
UP000007266
+ More
UP000235965 UP000095301 UP000095300 UP000192223 UP000000803 UP000008792 UP000008711 UP000192221 UP000009192 UP000075880 UP000183832 UP000007801 UP000002282 UP000268350 UP000001819 UP000002320 UP000007798 UP000007062 UP000075881 UP000075885 UP000008820 UP000001070 UP000075900 UP000076407 UP000075902 UP000075884 UP000075840 UP000245037 UP000075882 UP000075903 UP000092553 UP000015102 UP000009046 UP000215335 UP000002358 UP000092461
UP000235965 UP000095301 UP000095300 UP000192223 UP000000803 UP000008792 UP000008711 UP000192221 UP000009192 UP000075880 UP000183832 UP000007801 UP000002282 UP000268350 UP000001819 UP000002320 UP000007798 UP000007062 UP000075881 UP000075885 UP000008820 UP000001070 UP000075900 UP000076407 UP000075902 UP000075884 UP000075840 UP000245037 UP000075882 UP000075903 UP000092553 UP000015102 UP000009046 UP000215335 UP000002358 UP000092461
PRIDE
Pfam
PF00003 7tm_3
ProteinModelPortal
H9J7J0
A0A3S2L4C5
A0A2H1W525
A0A194PS45
A0A212EL71
A0A194QU36
+ More
I4DMA5 D6WE24 A0A2J7RBE3 A0A2J7RBE7 A0A1Y1MUP5 A0A1I8N4A0 A0A1I8N484 A0A1I8N4A1 A0A1I8Q7S6 A0A1I8Q7S3 A0A1W4X0S3 A0A0J9S0Z4 M9PIJ4 A0A0Q9WKP1 A0A0Q5U5X1 A0A1W4UE79 A0A0Q9XE83 A0A182JLV2 A0A1J1J762 A0A0P8XT72 B4LD89 A0A0R1DYN0 B4KUW8 A0A3B0J3E4 A0A0R3P3X3 A0A1W4WZR5 A0A336MIP6 A0A336L767 A0A0A9YWZ7 A0A0Q9WSK3 A0A0Q9XMS6 B0WM52 A0A0Q9WP33 A0A1B6JFN1 A0A1B6LNM2 Q5TMI1 A0A182KF49 A0A023F3D5 A0A182PWD9 A0A1B6LD82 A0A1B6FZX9 Q17N19 A0A1B6GUY7 B4IYU8 A0A182RQM6 A0A182WX88 A0A182U573 A0A182N2F0 A0A182HN15 A0A2P8ZIZ8 A0A182LNT5 A0A182V7C9 A0A0M5J8R4 A0A0R1DYP2 B4PDI2 T1GGZ3 E0VHU6 A0A232FFR2 K7JHE1 A0A1B0CM31
I4DMA5 D6WE24 A0A2J7RBE3 A0A2J7RBE7 A0A1Y1MUP5 A0A1I8N4A0 A0A1I8N484 A0A1I8N4A1 A0A1I8Q7S6 A0A1I8Q7S3 A0A1W4X0S3 A0A0J9S0Z4 M9PIJ4 A0A0Q9WKP1 A0A0Q5U5X1 A0A1W4UE79 A0A0Q9XE83 A0A182JLV2 A0A1J1J762 A0A0P8XT72 B4LD89 A0A0R1DYN0 B4KUW8 A0A3B0J3E4 A0A0R3P3X3 A0A1W4WZR5 A0A336MIP6 A0A336L767 A0A0A9YWZ7 A0A0Q9WSK3 A0A0Q9XMS6 B0WM52 A0A0Q9WP33 A0A1B6JFN1 A0A1B6LNM2 Q5TMI1 A0A182KF49 A0A023F3D5 A0A182PWD9 A0A1B6LD82 A0A1B6FZX9 Q17N19 A0A1B6GUY7 B4IYU8 A0A182RQM6 A0A182WX88 A0A182U573 A0A182N2F0 A0A182HN15 A0A2P8ZIZ8 A0A182LNT5 A0A182V7C9 A0A0M5J8R4 A0A0R1DYP2 B4PDI2 T1GGZ3 E0VHU6 A0A232FFR2 K7JHE1 A0A1B0CM31
Ontologies
GO
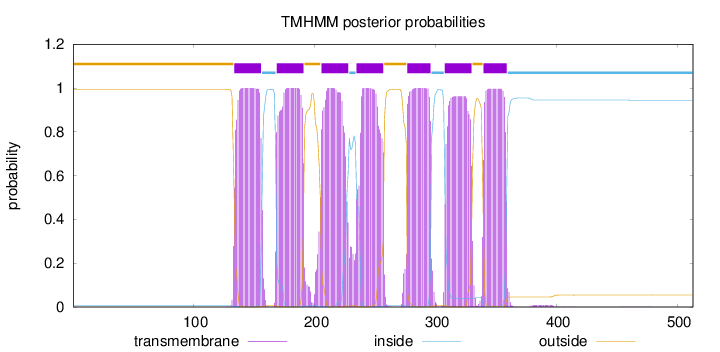
Topology
Length:
513
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
150.9693
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00594
outside
1 - 133
TMhelix
134 - 156
inside
157 - 168
TMhelix
169 - 191
outside
192 - 205
TMhelix
206 - 228
inside
229 - 234
TMhelix
235 - 257
outside
258 - 276
TMhelix
277 - 296
inside
297 - 307
TMhelix
308 - 330
outside
331 - 339
TMhelix
340 - 359
inside
360 - 513
Population Genetic Test Statistics
Pi
37.261954
Theta
62.353131
Tajima's D
-0.558668
CLR
0.547145
CSRT
0.229488525573721
Interpretation
Uncertain
